Transcriptome changes associated with anaerobic growth in Yersinia intermedia (ATCC29909)
- PMID: 24116118
- PMCID: PMC3792023
- DOI: 10.1371/journal.pone.0076567
Transcriptome changes associated with anaerobic growth in Yersinia intermedia (ATCC29909)
Abstract
Background: The yersiniae (Enterobacteriaceae) occupy a variety of niches, including some in human and flea hosts. Metabolic adaptations of the yersiniae, which contribute to their success in these specialized environments, remain largely unknown. We report results of an investigation of the transcriptome under aerobic and anaerobic conditions for Y. intermedia, a non-pathogenic member of the genus that has been used as a research surrogate for Y. pestis. Y. intermedia shares characteristics of pathogenic yersiniae, but is not known to cause disease in humans. Oxygen restriction is an important environmental stimulus experienced by many bacteria during their life-cycles and greatly influences their survival in specific environments. How oxygen availability affects physiology in the yersiniae is of importance in their life cycles but has not been extensively characterized.
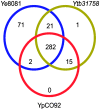
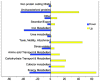
Methodology/principal findings: Tiled oligonucleotide arrays based on a draft genome sequence of Y. intermedia were used in transcript profiling experiments to identify genes that change expression in response to oxygen availability during growth in minimal media with glucose. The expression of more than 400 genes, constituting about 10% of the genome, was significantly altered due to oxygen-limitation in early log phase under these conditions. Broad functional categorization indicated that, in addition to genes involved in central metabolism, genes involved in adaptation to stress and genes likely involved with host interactions were affected by oxygen-availability. Notable among these, were genes encoding functions for motility, chemotaxis and biosynthesis of cobalamin, which were up-regulated and those for iron/heme utilization, methionine metabolism and urease, which were down-regulated.
Conclusions/significance: This is the first transcriptome analysis of a non-pathogenic Yersinia spp. and one of few elucidating the global response to oxygen limitation for any of the yersiniae. Thus this study lays the foundation for further experimental characterization of oxygen-responsive genes and pathways in this ecologically diverse genus.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

