Hybrid adeno-associated viral vectors utilizing transposase-mediated somatic integration for stable transgene expression in human cells
- PMID: 24116154
- PMCID: PMC3792901
- DOI: 10.1371/journal.pone.0076771
Hybrid adeno-associated viral vectors utilizing transposase-mediated somatic integration for stable transgene expression in human cells
Erratum in
-
Correction: Hybrid Adeno-Associated Viral Vectors Utilizing Transposase-Mediated Somatic Integration for Stable Transgene Expression in Human Cells.PLoS One. 2020 Jan 30;15(1):e0228707. doi: 10.1371/journal.pone.0228707. eCollection 2020. PLoS One. 2020. PMID: 31999798 Free PMC article.
Abstract
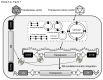
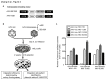
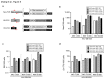
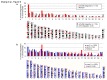
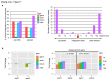
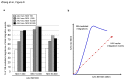
Recombinant adeno-associated viral (AAV) vectors have been shown to be one of the most promising vectors for therapeutic gene delivery because they can induce efficient and long-term transduction in non-dividing cells with negligible side-effects. However, as AAV vectors mostly remain episomal, vector genomes and transgene expression are lost in dividing cells. Therefore, to stably transduce cells, we developed a novel AAV/transposase hybrid-vector. To facilitate SB-mediated transposition from the rAAV genome, we established a system in which one AAV vector contains the transposon with the gene of interest and the second vector delivers the hyperactive Sleeping Beauty (SB) transposase SB100X. Human cells were infected with the AAV-transposon vector and the transposase was provided in trans either by transient and stable plasmid transfection or by AAV vector transduction. We found that groups which received the hyperactive transposase SB100X showed significantly increased colony forming numbers indicating enhanced integration efficiencies. Furthermore, we found that transgene copy numbers in transduced cells were dose-dependent and that predominantly SB transposase-mediated transposition contributed to stabilization of the transgene. Based on a plasmid rescue strategy and a linear-amplification mediated PCR (LAM-PCR) protocol we analysed the SB100X-mediated integration profile after transposition from the AAV vector. A total of 1840 integration events were identified which revealed a close to random integration profile. In summary, we show for the first time that AAV vectors can serve as template for SB transposase mediated somatic integration. We developed the first prototype of this hybrid-vector system which with further improvements may be explored for treatment of diseases which originate from rapidly dividing cells.
Conflict of interest statement
Figures


Similar articles
-
Integration profile and safety of an adenovirus hybrid-vector utilizing hyperactive sleeping beauty transposase for somatic integration.PLoS One. 2013 Oct 4;8(10):e75344. doi: 10.1371/journal.pone.0075344. eCollection 2013. PLoS One. 2013. PMID: 24124483 Free PMC article.
-
Stable transduction of the neonatal mouse liver using a hybrid rAAV/sleeping beauty transposon gene delivery system.J Gene Med. 2024 Aug;26(8):e3726. doi: 10.1002/jgm.3726. J Gene Med. 2024. PMID: 39160647
-
Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells.Mol Ther. 2011 Aug;19(8):1499-510. doi: 10.1038/mt.2011.47. Epub 2011 Apr 5. Mol Ther. 2011. PMID: 21468003 Free PMC article.
-
Development of adenovirus hybrid vectors for Sleeping Beauty transposition in large mammals.Curr Gene Ther. 2011 Oct;11(5):363-74. doi: 10.2174/156652311797415890. Curr Gene Ther. 2011. PMID: 21888620 Review.
-
Adeno-associated virus vector integration.Curr Opin Mol Ther. 2009 Aug;11(4):442-7. Curr Opin Mol Ther. 2009. PMID: 19649989 Free PMC article. Review.
Cited by
-
The best of both worlds: AAV-mediated gene transfer empowered by LNP delivery of Sleeping Beauty transposase for durable transgene expression in vivo.Mol Ther. 2024 Oct 2;32(10):3211-3214. doi: 10.1016/j.ymthe.2024.09.002. Epub 2024 Sep 25. Mol Ther. 2024. PMID: 39326408 No abstract available.
-
Genomic analysis of Sleeping Beauty transposon integration in human somatic cells.PLoS One. 2014 Nov 12;9(11):e112712. doi: 10.1371/journal.pone.0112712. eCollection 2014. PLoS One. 2014. PMID: 25390293 Free PMC article.
-
A High-Capacity Adenoviral Hybrid Vector System Utilizing the Hyperactive Sleeping Beauty Transposase SB100X for Enhanced Integration.Mol Ther Nucleic Acids. 2016 Jul 19;5(7):e337. doi: 10.1038/mtna.2016.44. Mol Ther Nucleic Acids. 2016. PMID: 27434682 Free PMC article.
-
Driving DNA transposition by lentiviral protein transduction.Mob Genet Elements. 2014 Jun 23;4:e29591. doi: 10.4161/mge.29591. eCollection 2014. Mob Genet Elements. 2014. PMID: 25057443 Free PMC article.
-
Correction: Hybrid Adeno-Associated Viral Vectors Utilizing Transposase-Mediated Somatic Integration for Stable Transgene Expression in Human Cells.PLoS One. 2020 Jan 30;15(1):e0228707. doi: 10.1371/journal.pone.0228707. eCollection 2020. PLoS One. 2020. PMID: 31999798 Free PMC article.
References
-
- Wiley (2013) Gene Therapy Clinical Trials Worldwide. J Gene Med.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

