A DNA-based semantic fusion model for remote sensing data
- PMID: 24116207
- PMCID: PMC3792926
- DOI: 10.1371/journal.pone.0077090
A DNA-based semantic fusion model for remote sensing data
Abstract
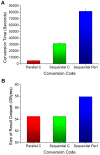
Semantic technology plays a key role in various domains, from conversation understanding to algorithm analysis. As the most efficient semantic tool, ontology can represent, process and manage the widespread knowledge. Nowadays, many researchers use ontology to collect and organize data's semantic information in order to maximize research productivity. In this paper, we firstly describe our work on the development of a remote sensing data ontology, with a primary focus on semantic fusion-driven research for big data. Our ontology is made up of 1,264 concepts and 2,030 semantic relationships. However, the growth of big data is straining the capacities of current semantic fusion and reasoning practices. Considering the massive parallelism of DNA strands, we propose a novel DNA-based semantic fusion model. In this model, a parallel strategy is developed to encode the semantic information in DNA for a large volume of remote sensing data. The semantic information is read in a parallel and bit-wise manner and an individual bit is converted to a base. By doing so, a considerable amount of conversion time can be saved, i.e., the cluster-based multi-processes program can reduce the conversion time from 81,536 seconds to 4,937 seconds for 4.34 GB source data files. Moreover, the size of result file recording DNA sequences is 54.51 GB for parallel C program compared with 57.89 GB for sequential Perl. This shows that our parallel method can also reduce the DNA synthesis cost. In addition, data types are encoded in our model, which is a basis for building type system in our future DNA computer. Finally, we describe theoretically an algorithm for DNA-based semantic fusion. This algorithm enables the process of integration of the knowledge from disparate remote sensing data sources into a consistent, accurate, and complete representation. This process depends solely on ligation reaction and screening operations instead of the ontology.
Conflict of interest statement
Figures






Similar articles
-
A fuzzy-ontology-oriented case-based reasoning framework for semantic diabetes diagnosis.Artif Intell Med. 2015 Nov;65(3):179-208. doi: 10.1016/j.artmed.2015.08.003. Epub 2015 Aug 14. Artif Intell Med. 2015. PMID: 26303105
-
Aggregating the syntactic and semantic similarity of healthcare data towards their transformation to HL7 FHIR through ontology matching.Int J Med Inform. 2019 Dec;132:104002. doi: 10.1016/j.ijmedinf.2019.104002. Epub 2019 Oct 5. Int J Med Inform. 2019. PMID: 31629311
-
Using ontology-based semantic similarity to facilitate the article screening process for systematic reviews.J Biomed Inform. 2017 May;69:33-42. doi: 10.1016/j.jbi.2017.03.007. Epub 2017 Mar 14. J Biomed Inform. 2017. PMID: 28302519
-
KaBOB: ontology-based semantic integration of biomedical databases.BMC Bioinformatics. 2015 Apr 23;16(1):126. doi: 10.1186/s12859-015-0559-3. BMC Bioinformatics. 2015. PMID: 25903923 Free PMC article.
-
Semantic Data Mining in Ubiquitous Sensing: A Survey.Sensors (Basel). 2021 Jun 24;21(13):4322. doi: 10.3390/s21134322. Sensors (Basel). 2021. PMID: 34202654 Free PMC article. Review.
References
-
- Adleman LM (1994) Molecular computation of solutions to combinatorial problems. Science 266: 1021–1024. - PubMed
-
- Lipton R (1995) DNA solution of hard computational problems. Science 268: 542–545. - PubMed
-
- Bancroft C, Bowler T, Bloom B, Clelland CT (2001) Long-term storage of information in DNA. Science 293: 1763–1765. - PubMed
-
- Renear A, Palmer C (2009) Strategic reading, ontologies, and the future of scientific publishing. Science 325: 828–832. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

