Role and regulation of heme iron acquisition in gram-negative pathogens
- PMID: 24116354
- PMCID: PMC3792355
- DOI: 10.3389/fcimb.2013.00055
Role and regulation of heme iron acquisition in gram-negative pathogens
Abstract
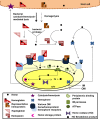
Bacteria that reside in animal tissues and/or cells must acquire iron from their host. However, almost all of the host iron is sequestered in iron-containing compounds and proteins, the majority of which is found within heme molecules. Thus, likely iron sources for bacterial pathogens (and non-pathogenic symbionts) are free heme and heme-containing proteins. Furthermore, the cellular location of the bacterial within the host (intra or extracellular) influences the amount and nature of the iron containing compounds available for transport. The low level of free iron in the host, coupled with the presence of numerous different heme sources, has resulted in a wide range of high-affinity iron acquisition strategies within bacteria. However, since excess iron and heme are toxic to bacteria, expression of these acquisition systems is highly regulated. Precise expression in the correct host environment at the appropriate times enables heme iron acquisitions systems to contribute to the growth of bacterial pathogens within the host. This mini-review will highlight some of the recent findings in these areas for gram-negative pathogens.
Keywords: Fur; hem; heme; hemin; hemoglobin; iron; pathogens; regulation.
Figures
Similar articles
-
Heme Uptake and Utilization by Gram-Negative Bacterial Pathogens.Front Cell Infect Microbiol. 2019 Mar 29;9:81. doi: 10.3389/fcimb.2019.00081. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 30984629 Free PMC article. Review.
-
Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores.Curr Opin Microbiol. 2000 Apr;3(2):215-20. doi: 10.1016/s1369-5274(00)00078-3. Curr Opin Microbiol. 2000. PMID: 10744995 Review.
-
[Mechanisms and regulation of iron and heme utilization in Gram-negative bacteria].Postepy Biochem. 2005;51(2):198-208. Postepy Biochem. 2005. PMID: 16209357 Review. Polish.
-
HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae.J Bacteriol. 2009 Apr;191(8):2638-48. doi: 10.1128/JB.01784-08. Epub 2009 Feb 6. J Bacteriol. 2009. PMID: 19201805 Free PMC article.
-
Heme Synthesis and Acquisition in Bacterial Pathogens.J Mol Biol. 2016 Aug 28;428(17):3408-28. doi: 10.1016/j.jmb.2016.03.018. Epub 2016 Mar 24. J Mol Biol. 2016. PMID: 27019298 Free PMC article. Review.
Cited by
-
Bacteroides thetaiotaomicron, a Model Gastrointestinal Tract Species, Prefers Heme as an Iron Source, Yields Protoporphyrin IX as a Product, and Acts as a Heme Reservoir.Microbiol Spectr. 2023 Mar 2;11(2):e0481522. doi: 10.1128/spectrum.04815-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36862015 Free PMC article.
-
The transcriptional regulator IscR integrates host-derived nitrosative stress and iron starvation in activation of the vvhBA operon in Vibrio vulnificus.J Biol Chem. 2020 Apr 17;295(16):5350-5361. doi: 10.1074/jbc.RA120.012724. Epub 2020 Mar 13. J Biol Chem. 2020. PMID: 32169898 Free PMC article.
-
TonB-Dependent Heme/Hemoglobin Utilization by Caulobacter crescentus HutA.J Bacteriol. 2017 Feb 28;199(6):e00723-16. doi: 10.1128/JB.00723-16. Print 2017 Mar 15. J Bacteriol. 2017. PMID: 28031282 Free PMC article.
-
The intricacies of Acinetobacter baumannii: a multifaceted comprehensive review of a multidrug-resistant pathogen and its clinical significance and implications.Front Microbiol. 2025 May 12;16:1565965. doi: 10.3389/fmicb.2025.1565965. eCollection 2025. Front Microbiol. 2025. PMID: 40444001 Free PMC article. Review.
-
Dual Inhibition of Klebsiella pneumoniae and Pseudomonas aeruginosa Iron Metabolism Using Gallium Porphyrin and Gallium Nitrate.ACS Infect Dis. 2019 Sep 13;5(9):1559-1569. doi: 10.1021/acsinfecdis.9b00100. Epub 2019 Jul 16. ACS Infect Dis. 2019. PMID: 31264851 Free PMC article.
References
-
- Aduse-Opoku J., Slaney J. M., Rangarajan M., Muir J., Young K. A., Curtis M. A. (1997). The Tla protein of Porphyromonas gingivalis W50: a homolog of the RI protease precursor (PrpRI) is an outer membrane receptor required for growth on low levels of hemin. J. Bacteriol. 179, 4778–4788 - PMC - PubMed
-
- Antunes L. C., Imperi F., Towner K. J., Visca P. (2011). Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162, 279–284 10.1016/j.resmic.2010.10.010 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

