Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology
- PMID: 24116388
- PMCID: PMC3837439
- DOI: 10.1089/omi.2013.0017
Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology
Abstract
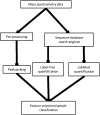
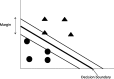
Mass spectrometry is an analytical technique for the characterization of biological samples and is increasingly used in omics studies because of its targeted, nontargeted, and high throughput abilities. However, due to the large datasets generated, it requires informatics approaches such as machine learning techniques to analyze and interpret relevant data. Machine learning can be applied to MS-derived proteomics data in two ways. First, directly to mass spectral peaks and second, to proteins identified by sequence database searching, although relative protein quantification is required for the latter. Machine learning has been applied to mass spectrometry data from different biological disciplines, particularly for various cancers. The aims of such investigations have been to identify biomarkers and to aid in diagnosis, prognosis, and treatment of specific diseases. This review describes how machine learning has been applied to proteomics tandem mass spectrometry data. This includes how it can be used to identify proteins suitable for use as biomarkers of disease and for classification of samples into disease or treatment groups, which may be applicable for diagnostics. It also includes the challenges faced by such investigations, such as prediction of proteins present, protein quantification, planning for the use of machine learning, and small sample sizes.
Figures



References
-
- Abeel T. Helleputte T. Van De Peer Y. Dupont P. Saeys Y. Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics. 2010;26:392–398. - PubMed
-
- Adam BL. Qu Y. Davis JW, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. - PubMed
-
- Aebersold R. Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
