Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle
- PMID: 24119401
- PMCID: PMC4039290
- DOI: 10.1016/j.molcel.2013.09.009
Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle
Abstract
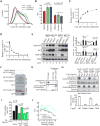
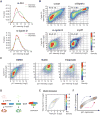
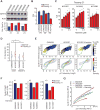
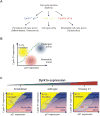
Mammalian cells have a remarkable capacity to compensate for heterozygous gene loss or extra gene copies. One exception is Down syndrome (DS), where a third copy of chromosome 21 mediates neurogenesis defects and lowers the frequency of solid tumors. Here we combine live-cell imaging and single-cell analysis to show that increased dosage of chromosome 21-localized Dyrk1a steeply increases G1 cell cycle duration through direct phosphorylation and degradation of cyclin D1 (CycD1). DS-derived fibroblasts showed analogous cell cycle changes that were reversed by Dyrk1a inhibition. Furthermore, reducing Dyrk1a activity increased CycD1 expression to force a bifurcation, with one subpopulation of cells accelerating proliferation and the other arresting proliferation by costabilizing CycD1 and the CDK inhibitor p21. Thus, dosage of Dyrk1a repositions cells within a p21-CycD1 signaling map, directing each cell to either proliferate or to follow two distinct cell cycle exit pathways characterized by high or low CycD1 and p21 levels.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Altafaj X, Dierssen M, Baamonde C, Marti E, Visa J, Guimera J, Oset M, Gonzalez JR, Florez J, Fillat C, et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum Mol Genet. 2001;10:1915–1923. - PubMed
-
- Becker L, Mito T, Takashima S, Onodera K. Growth and development of the brain in Down syndrome. Prog Clin Biol Res. 1991;373:133–152. - PubMed
-
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

