Intratumoral genetic heterogeneity in metastatic melanoma is accompanied by variation in malignant behaviors
- PMID: 24119551
- PMCID: PMC3852494
- DOI: 10.1186/1755-8794-6-40
Intratumoral genetic heterogeneity in metastatic melanoma is accompanied by variation in malignant behaviors
Abstract
Background: Intratumoral heterogeneity is a major obstacle for the treatment of cancer, as the presence of even minor populations that are insensitive to therapy can lead to disease relapse. Increased clonal diversity has been correlated with a poor prognosis for cancer patients, and we therefore examined genetic, transcriptional, and functional diversity in metastatic melanoma.
Methods: Amplicon sequencing and SNP microarrays were used to profile somatic mutations and DNA copy number changes in multiple regions from metastatic lesions. Clonal genetic and transcriptional heterogeneity was also assessed in single cell clones from early passage cell lines, which were then subjected to clonogenicity and drug sensitivity assays.
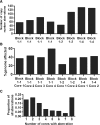
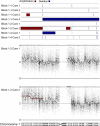
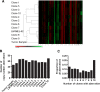
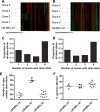
Results: MAPK pathway and tumor suppressor mutations were identified in all regions of the melanoma metastases analyzed. In contrast, we identified copy number abnormalities present in only some regions in addition to homogeneously present changes, suggesting ongoing genetic evolution following metastatic spread. Copy number heterogeneity from a tumor was represented in matched cell line clones, which also varied in their clonogenicity and drug sensitivity. Minor clones were identified based on dissimilarity to the parental cell line, and these clones were the most clonogenic and least sensitive to drugs. Finally, treatment of a polyclonal cell line with paclitaxel to enrich for drug-resistant cells resulted in the adoption of a gene expression profile with features of one of the minor clones, supporting the idea that these populations can mediate disease relapse.
Conclusion: Our results support the hypothesis that minor clones might have major consequences for patient outcomes in melanoma.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

