Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis
- PMID: 24120662
- PMCID: PMC4691852
- DOI: 10.1016/j.molcel.2013.09.006
Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis
Abstract
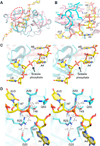
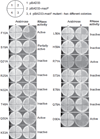
MazF is an mRNA interferase, which, upon activation during stress conditions, cleaves mRNAs in a sequence-specific manner, resulting in cellular growth arrest. During normal growth conditions, the MazF toxin is inactivated through binding to its cognate antitoxin, MazE. How MazF specifically recognizes its mRNA target and carries out cleavage and how the formation of the MazE-MazF complex inactivates MazF remain unclear. We present crystal structures of MazF in complex with mRNA substrate and antitoxin MazE in Bacillus subtilis. The structure of MazF in complex with uncleavable UUdUACAUAA RNA substrate defines the molecular basis underlying the sequence-specific recognition of UACAU and the role of residues involved in the cleavage through site-specific mutational studies. The structure of the heterohexameric (MazF)2-(MazE)2-(MazF)2 complex in Bacillus subtilis, supplemented by mutational data, demonstrates that the positioning of the C-terminal helical segment of MazE within the RNA-binding channel of the MazF dimer prevents mRNA binding and cleavage by MazF.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


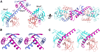


References
-
- Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 2005;30:672–679. - PubMed
-
- De Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, Wyns L, De Greve H, Loris R. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell. 2009;35:154–163. - PubMed
-
- Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

