A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation
- PMID: 24127489
- PMCID: PMC3804941
- DOI: 10.1084/jem.20122709
A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation
Abstract
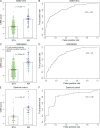
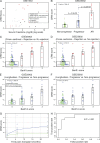
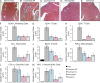
Using meta-analysis of eight independent transplant datasets (236 graft biopsy samples) from four organs, we identified a common rejection module (CRM) consisting of 11 genes that were significantly overexpressed in acute rejection (AR) across all transplanted organs. The CRM genes could diagnose AR with high specificity and sensitivity in three additional independent cohorts (794 samples). In another two independent cohorts (151 renal transplant biopsies), the CRM genes correlated with the extent of graft injury and predicted future injury to a graft using protocol biopsies. Inferred drug mechanisms from the literature suggested that two FDA-approved drugs (atorvastatin and dasatinib), approved for nontransplant indications, could regulate specific CRM genes and reduce the number of graft-infiltrating cells during AR. We treated mice with HLA-mismatched mouse cardiac transplant with atorvastatin and dasatinib and showed reduction of the CRM genes, significant reduction of graft-infiltrating cells, and extended graft survival. We further validated the beneficial effect of atorvastatin on graft survival by retrospective analysis of electronic medical records of a single-center cohort of 2,515 renal transplant patients followed for up to 22 yr. In conclusion, we identified a CRM in transplantation that provides new opportunities for diagnosis, drug repositioning, and rational drug design.
Figures




References
-
- Baron U., Floess S., Wieczorek G., Baumann K., Grützkau A., Dong J., Thiel A., Boeld T.J., Hoffmann P., Edinger M., et al. 2007. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur. J. Immunol. 37:2378–2389 10.1002/eji.200737594 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

