Structural destabilization of DNA duplexes containing single-base lesions investigated by nanopore measurements
- PMID: 24128275
- PMCID: PMC3867232
- DOI: 10.1021/bi4009825
Structural destabilization of DNA duplexes containing single-base lesions investigated by nanopore measurements
Abstract
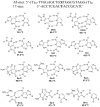
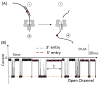
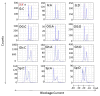
The influence of DNA duplex structural destabilization introduced by a single base-pair modification was investigated by nanopore measurements. A series of 11 modified base pairs were introduced into the context of an otherwise complementary DNA duplex formed by a 17-mer and a 65-mer such that the overhanging ends comprised poly(dT)23 tails, generating a representative set of duplexes that display a range of unzipping mechanistic behaviors and kinetic stabilities. The guanine oxidation products 8-oxo-7,8-dihydroguanine (OG), guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp) were paired with either cytosine (C), adenine (A), or 2,6-diaminopurine (D) to form modified base pairs. The mechanism and kinetic rate constants of duplex dissociation were determined by threading either the 3' or 5' overhangs into an α-hemolysin (α-HL) channel under an electrical field and measuring the distributions of unzipping times at constant force. In order of decreasing thermodynamic stability (as measured by duplex melting points), the rate of duplex dissociation increases, and the mechanism evolves from a first-order reaction to two sequential first-order reactions. These measurements allow us to rank the kinetic stability of lesion-containing duplexes relative to the canonical G:C base pair in which the OG:C, Gh:C, and Sp:C base pairs are, respectively, 3-200 times less stable. The rate constants also depend on whether unzipping was initiated from the 3' versus 5' side of the duplex. The kinetic stability of these duplexes was interpreted in terms of the structural destabilization introduced by the single base-pair modification. Specifically, a large distortion of the duplex backbone introduced by the presence of the highly oxidized guanine products Sp and Gh leads to a rapid two-step unzipping. The number of hydrogen bonds in the modified base pair plays a lesser role in determining the kinetics of duplex dissociation.
Figures

Similar articles
-
Unzipping kinetics of duplex DNA containing oxidized lesions in an α-hemolysin nanopore.J Am Chem Soc. 2012 Jul 4;134(26):11006-11. doi: 10.1021/ja304169n. Epub 2012 Jun 25. J Am Chem Soc. 2012. PMID: 22690806 Free PMC article.
-
Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2'-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing.J Am Chem Soc. 2012 Sep 12;134(36):15091-102. doi: 10.1021/ja306077b. Epub 2012 Aug 29. J Am Chem Soc. 2012. PMID: 22880947 Free PMC article.
-
Sequence-specific single-molecule analysis of 8-oxo-7,8-dihydroguanine lesions in DNA based on unzipping kinetics of complementary probes in ion channel recordings.J Am Chem Soc. 2011 Sep 21;133(37):14778-84. doi: 10.1021/ja205653v. Epub 2011 Aug 29. J Am Chem Soc. 2011. PMID: 21875081 Free PMC article.
-
Unusual structural features of hydantoin lesions translate into efficient recognition by Escherichia coli Fpg.Biochemistry. 2007 Aug 21;46(33):9355-65. doi: 10.1021/bi602459v. Epub 2007 Jul 27. Biochemistry. 2007. PMID: 17655276 Free PMC article.
-
Nanopore Analysis of the 5-Guanidinohydantoin to Iminoallantoin Isomerization in Duplex DNA.J Org Chem. 2018 Apr 6;83(7):3973-3978. doi: 10.1021/acs.joc.8b00317. Epub 2018 Mar 8. J Org Chem. 2018. PMID: 29490132 Free PMC article.
Cited by
-
RNA Editing of the Human DNA Glycosylase NEIL1 Alters Its Removal of 5-Hydroxyuracil Lesions in DNA.Biochemistry. 2021 May 18;60(19):1485-1497. doi: 10.1021/acs.biochem.1c00062. Epub 2021 Apr 30. Biochemistry. 2021. PMID: 33929180 Free PMC article.
-
Removal of oxidatively generated DNA damage by overlapping repair pathways.Free Radic Biol Med. 2017 Jun;107:53-61. doi: 10.1016/j.freeradbiomed.2016.10.507. Epub 2016 Nov 4. Free Radic Biol Med. 2017. PMID: 27818219 Free PMC article. Review.
-
The shaping of a molecular linguist: How a career studying DNA energetics revealed the language of molecular communication.J Biol Chem. 2021 Jan-Jun;296:100522. doi: 10.1016/j.jbc.2021.100522. Epub 2021 Apr 7. J Biol Chem. 2021. PMID: 34237886 Free PMC article.
-
Unzipping of A-Form DNA-RNA, A-Form DNA-PNA, and B-Form DNA-DNA in the α-Hemolysin Nanopore.Biophys J. 2016 Jan 19;110(2):306-314. doi: 10.1016/j.bpj.2015.11.020. Biophys J. 2016. PMID: 26789754 Free PMC article.
-
Base and Nucleotide Excision Repair of Oxidatively Generated Guanine Lesions in DNA.J Biol Chem. 2016 Mar 4;291(10):5309-19. doi: 10.1074/jbc.M115.693218. Epub 2016 Jan 5. J Biol Chem. 2016. PMID: 26733197 Free PMC article.
References
-
- Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

