Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit
- PMID: 24129118
- PMCID: PMC7113674
- DOI: 10.1016/j.antiviral.2013.10.002
Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit
Abstract
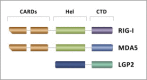
Double-stranded RNA (dsRNA) is synthesized during the course of infection by RNA viruses as a byproduct of replication and transcription and acts as a potent trigger of the host innate antiviral response. In the cytoplasm of the infected cell, recognition of the presence of viral dsRNA as a signature of "non-self" nucleic acid is carried out by RIG-I-like receptors (RLRs), a set of dedicated helicases whose activation leads to the production of type I interferon α/β (IFN-α/β). To overcome the innate antiviral response, RNA viruses encode suppressors of IFN-α/β induction, which block RLRs recognition of dsRNA by means of different mechanisms that can be categorized into: (i) dsRNA binding and/or shielding ("hide"), (ii) dsRNA termini processing ("mask") and (iii) direct interaction with components of the RLRs pathway ("hit"). In light of recent functional, biochemical and structural findings, we review the inhibition mechanisms of RLRs recognition of dsRNA displayed by a number of highly pathogenic RNA viruses with different disease phenotypes such as haemorrhagic fever (Ebola, Marburg, Lassa fever, Lujo, Machupo, Junin, Guanarito, Crimean-Congo, Rift Valley fever, dengue), severe respiratory disease (influenza, SARS, Hendra, Hantaan, Sin Nombre, Andes) and encephalitis (Nipah, West Nile).
Keywords: Innate immune system evasion; Interferon alpha/beta; LGP2; MDA5; RIG-I; Viral dsRNA detection.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells.EMBO J. 2018 Feb 15;37(4):e97479. doi: 10.15252/embj.201797479. Epub 2018 Jan 19. EMBO J. 2018. PMID: 29351913 Free PMC article.
-
[Recent progress of the mechanisms for RNA viruses to block the recognition of dsRNA with RIG-I-like receptors].Bing Du Xue Bao. 2014 Nov;30(6):704-12. Bing Du Xue Bao. 2014. PMID: 25868287 Review. Chinese.
-
Innate immunity to virus infection.Immunol Rev. 2009 Jan;227(1):75-86. doi: 10.1111/j.1600-065X.2008.00737.x. Immunol Rev. 2009. PMID: 19120477 Free PMC article. Review.
-
Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses.Nature. 2006 May 4;441(7089):101-5. doi: 10.1038/nature04734. Epub 2006 Apr 9. Nature. 2006. PMID: 16625202
-
Processing of genome 5' termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction.PLoS One. 2008 Apr 30;3(4):e2032. doi: 10.1371/journal.pone.0002032. PLoS One. 2008. PMID: 18446221 Free PMC article.
Cited by
-
Innate Immune Evasion by Human Respiratory RNA Viruses.J Innate Immun. 2020;12(1):4-20. doi: 10.1159/000503030. Epub 2019 Oct 14. J Innate Immun. 2020. PMID: 31610541 Free PMC article. Review.
-
Structural insights into the Middle East respiratory syndrome coronavirus 4a protein and its dsRNA binding mechanism.Sci Rep. 2017 Sep 12;7(1):11362. doi: 10.1038/s41598-017-11736-6. Sci Rep. 2017. PMID: 28900197 Free PMC article.
-
Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein.Cell Host Microbe. 2015 Sep 9;18(3):333-44. doi: 10.1016/j.chom.2015.07.015. Epub 2015 Aug 27. Cell Host Microbe. 2015. PMID: 26320998 Free PMC article.
-
LINE-1 ORF1p Mimics Viral Innate Immune Evasion Mechanisms in Pancreatic Ductal Adenocarcinoma.Cancer Discov. 2025 May 2;15(5):1063-1082. doi: 10.1158/2159-8290.CD-24-1317. Cancer Discov. 2025. PMID: 39919290 Free PMC article.
-
Role of the early secretory pathway in SARS-CoV-2 infection.J Cell Biol. 2020 Sep 7;219(9):e202006005. doi: 10.1083/jcb.202006005. J Cell Biol. 2020. PMID: 32725137 Free PMC article. Review.
References
-
- Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D., Maringer K., Bernal-Rubio D., Shabman R.S., Simon V., Rodriguez-Madoz J.R., Mulder L.C.F., Barber G.N., Fernandez-Sesma A. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. - PMC - PubMed
-
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

