CXCR3/CCR5 pathways in metastatic melanoma patients treated with adoptive therapy and interleukin-2
- PMID: 24129241
- PMCID: PMC3817317
- DOI: 10.1038/bjc.2013.557
CXCR3/CCR5 pathways in metastatic melanoma patients treated with adoptive therapy and interleukin-2
Abstract
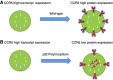
Background: Adoptive therapy with tumour-infiltrating lymphocytes (TILs) induces durable complete responses (CR) in ∼20% of patients with metastatic melanoma. The recruitment of T cells through CXCR3/CCR5 chemokine ligands is critical for immune-mediated rejection. We postulated that polymorphisms and/or expression of CXCR3/CCR5 in TILs and the expression of their ligands in tumour influence the migration of TILs to tumours and tumour regression.
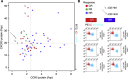
Methods: Tumour-infiltrating lymphocytes from 142 metastatic melanoma patients enrolled in adoptive therapy trials were genotyped for CXCR3 rs2280964 and CCR5-Δ32 deletion, which encodes a protein not expressed on the cell surface. Expression of CXCR3/CCR5 in TILs and CXCR3/CCR5 and ligand genes in 113 available parental tumours was also assessed. Tumour-infiltrating lymphocyte data were validated by flow cytometry (N=50).
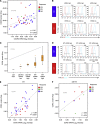
Results: The full gene expression/polymorphism model, which includes CXCR3 and CCR5 expression data, CCR5-Δ32 polymorphism data and their interaction, was significantly associated with both CR and overall response (OR; P=0.0009, and P=0.007, respectively). More in detail, the predicted underexpression of both CXCR3 and CCR5 according to gene expression and polymorphism data (protein prediction model, PPM) was associated with response to therapy (odds ratio=6.16 and 2.32, for CR and OR, respectively). Flow cytometric analysis confirmed the PPM. Coordinate upregulation of CXCL9, CXCL10, CXCL11, and CCL5 in pretreatment tumour biopsies was associated with OR.
Conclusion: Coordinate overexpression of CXCL9, CXCL10, CXCL11, and CCL5 in pretreatment tumours was associated with responsiveness to treatment. Conversely, CCR5-Δ32 polymorphism and CXCR3/CCR5 underexpression influence downregulation of the corresponding receptors in TILs and were associated with likelihood and degree of response.
Figures


Similar articles
-
T lymphocyte recruitment into renal cell carcinoma tissue: a role for chemokine receptors CXCR3, CXCR6, CCR5, and CCR6.Eur Urol. 2012 Feb;61(2):385-94. doi: 10.1016/j.eururo.2011.10.035. Epub 2011 Nov 4. Eur Urol. 2012. PMID: 22079021
-
Pro-inflammatory chemokine-chemokine receptor interactions within the Ewing sarcoma microenvironment determine CD8(+) T-lymphocyte infiltration and affect tumour progression.J Pathol. 2011 Feb;223(3):347-57. doi: 10.1002/path.2819. Epub 2010 Dec 10. J Pathol. 2011. PMID: 21171080
-
Infiltrating CD8+ T cells in oral lichen planus predominantly express CCR5 and CXCR3 and carry respective chemokine ligands RANTES/CCL5 and IP-10/CXCL10 in their cytolytic granules: a potential self-recruiting mechanism.Am J Pathol. 2003 Jul;163(1):261-8. doi: 10.1016/S0002-9440(10)63649-8. Am J Pathol. 2003. PMID: 12819030 Free PMC article.
-
CXCR3 Ligands in Cancer and Autoimmunity, Chemoattraction of Effector T Cells, and Beyond.Front Immunol. 2020 May 29;11:976. doi: 10.3389/fimmu.2020.00976. eCollection 2020. Front Immunol. 2020. PMID: 32547545 Free PMC article. Review.
-
Targeting of Th1-associated chemokine receptors CXCR3 and CCR5 as therapeutic strategy for inflammatory diseases.Mini Rev Med Chem. 2007 Nov;7(11):1089-96. doi: 10.2174/138955707782331768. Mini Rev Med Chem. 2007. PMID: 18045212 Review.
Cited by
-
Checkpoint Inhibitors and Their Application in Breast Cancer.Breast Care (Basel). 2016 Apr;11(2):108-15. doi: 10.1159/000445335. Epub 2016 Apr 26. Breast Care (Basel). 2016. PMID: 27239172 Free PMC article. Review.
-
Genetic Variation in CCL5 Signaling Genes and Triple Negative Breast Cancer: Susceptibility and Prognosis Implications.Front Oncol. 2019 Dec 6;9:1328. doi: 10.3389/fonc.2019.01328. eCollection 2019. Front Oncol. 2019. PMID: 31921621 Free PMC article.
-
A Perspective of Immunotherapy for Breast Cancer: Lessons Learned and Forward Directions for All Cancers.Breast Cancer (Auckl). 2015 Nov 2;9(Suppl 2):35-43. doi: 10.4137/BCBCR.S29425. eCollection 2015. Breast Cancer (Auckl). 2015. PMID: 26568682 Free PMC article. Review.
-
Cytotoxic T-Cell Trafficking Chemokine Profiles Correlate With Defined Mucosal Microbial Communities in Colorectal Cancer.Front Immunol. 2021 Sep 1;12:715559. doi: 10.3389/fimmu.2021.715559. eCollection 2021. Front Immunol. 2021. PMID: 34539647 Free PMC article.
-
HDAC inhibitors enhance the immunotherapy response of melanoma cells.Oncotarget. 2017 May 17;8(47):83155-83170. doi: 10.18632/oncotarget.17950. eCollection 2017 Oct 10. Oncotarget. 2017. PMID: 29137331 Free PMC article.
References
-
- Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, Dumur C, Wang E, Ramakrishnan V, Wang XY, Bear HD, Marincola FM, Manjili MH. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat. 2012;131 (3:871–880. - PMC - PubMed
-
- Bedognetti D, Balwit JM, Wang E, Disis ML, Britten CM, Delogu LG, Tomei S, Fox BA, Gajewski TF, Marincola FM, Butterfield LH. SITC/iSBTc cancer immunotherapy biomarkers resource document: online resources and useful tools—a compass in the land of biomarker discovery. J Transl Med. 2011;9:155. - PMC - PubMed
-
- Carrington M, Dean M, Martin MP, O′Brien SJ. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol Genet. 1999;8 (10:1939–1945. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

