SILAC-based phosphoproteomics reveals an inhibitory role of KSR1 in p53 transcriptional activity via modulation of DBC1
- PMID: 24129246
- PMCID: PMC3833216
- DOI: 10.1038/bjc.2013.628
SILAC-based phosphoproteomics reveals an inhibitory role of KSR1 in p53 transcriptional activity via modulation of DBC1
Abstract
Background: We have previously identified kinase suppressor of ras-1 (KSR1) as a potential regulatory gene in breast cancer. KSR1, originally described as a novel protein kinase, has a role in activation of mitogen-activated protein kinases. Emerging evidence has shown that KSR1 may have dual functions as an active kinase as well as a scaffold facilitating multiprotein complex assembly. Although efforts have been made to study the role of KSR1 in certain tumour types, its involvement in breast cancer remains unknown.
Methods: A quantitative mass spectrometry analysis using stable isotope labelling of amino acids in cell culture (SILAC) was implemented to identify KSR1-regulated phosphoproteins in breast cancer. In vitro luciferase assays, co-immunoprecipitation as well as western blotting experiments were performed to further study the function of KSR1 in breast cancer.
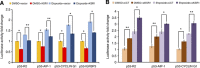
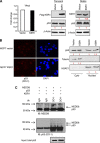
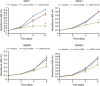
Results: Of significance, proteomic analysis reveals that KSR1 overexpression decreases deleted in breast cancer-1 (DBC1) phosphorylation. Furthermore, we show that KSR1 decreases the transcriptional activity of p53 by reducing the phosphorylation of DBC1, which leads to a reduced interaction of DBC1 with sirtuin-1 (SIRT1); this in turn enables SIRT1 to deacetylate p53.
Conclusion: Our findings integrate KSR1 into a network involving DBC1 and SIRT1, which results in the regulation of p53 acetylation and its transcriptional activity.
Figures



References
-
- Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001;93:1062–1074. - PubMed
-
- Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. - PubMed
-
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

