Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation
- PMID: 24129573
- PMCID: PMC3853261
- DOI: 10.1074/jbc.M113.511949
Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation
Abstract
Although lysine methylation is classically known to regulate histone function, its role in modulating antiviral restriction factor activity remains uncharacterized. Interferon-induced transmembrane protein 3 (IFITM3) was found monomethylated on its lysine 88 residue (IFITM3-K88me1) to reduce its antiviral activity, mediated by the lysine methyltransferase SET7. Vesicular stomatitis virus and influenza A virus infection increased IFITM3-K88me1 levels by promoting the interaction between IFITM3 and SET7, suggesting that this pathway could be hijacked to support infection; conversely, IFN-α reduced IFITM3-K88me1 levels. These findings may have important implications in the design of therapeutics targeting protein methylation against infectious diseases.
Keywords: Antiviral Agents; Antiviral Host Restriction Factors; Host Defense; Host-pathogen Interactions; IFITM3; Lysine Methylation; Post-translational Modification; Protein Methylation; SET7.
Figures


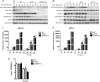
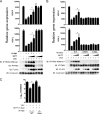


References
-
- Lewin A. R., Reid L. E., McMahon M., Stark G. R., Kerr I. M. (1991) Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199, 417–423 - PubMed
-
- Everitt A. R., Clare S., Pertel T., John S. P., Wash R. S., Smith S. E., Chin C. R., Feeley E. M., Sims J. S., Adams D. J., Wise H. M., Kane L., Goulding D., Digard P., Anttila V., Baillie J. K., Walsh T. S., Hume D. A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S. B., GenISIS Investigators, MOSAIC Investigators, Smyth R. L., Openshaw P. J., Dougan G., Brass A. L., Kellam P. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

