Analysis of functional redundancies within the Arabidopsis TCP transcription factor family
- PMID: 24129704
- PMCID: PMC3871820
- DOI: 10.1093/jxb/ert337
Analysis of functional redundancies within the Arabidopsis TCP transcription factor family
Abstract
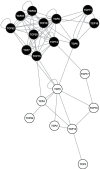
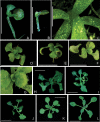
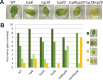
Analyses of the functions of TEOSINTE-LIKE1, CYCLOIDEA, and PROLIFERATING CELL FACTOR1 (TCP) transcription factors have been hampered by functional redundancy between its individual members. In general, putative functionally redundant genes are predicted based on sequence similarity and confirmed by genetic analysis. In the TCP family, however, identification is impeded by relatively low overall sequence similarity. In a search for functionally redundant TCP pairs that control Arabidopsis leaf development, this work performed an integrative bioinformatics analysis, combining protein sequence similarities, gene expression data, and results of pair-wise protein-protein interaction studies for the 24 members of the Arabidopsis TCP transcription factor family. For this, the work completed any lacking gene expression and protein-protein interaction data experimentally and then performed a comprehensive prediction of potential functional redundant TCP pairs. Subsequently, redundant functions could be confirmed for selected predicted TCP pairs by genetic and molecular analyses. It is demonstrated that the previously uncharacterized class I TCP19 gene plays a role in the control of leaf senescence in a redundant fashion with TCP20. Altogether, this work shows the power of combining classical genetic and molecular approaches with bioinformatics predictions to unravel functional redundancies in the TCP transcription factor family.
Keywords: Bioinformatics; TCP transcription factor.; gene regulation; leaf development; redundancy; senescence.
Figures


References
-
- Aoyama T, Chua N-H. 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. The Plant Journal 11, 605–612 - PubMed
-
- Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters 573, 83–92 - PubMed
-
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. 2006. Unequal genetic redundancies in Arabidopsis—a neglected phenomenon? Trends in Plant Science 11, 492–498 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

