Novel staphylococcal glycosyltransferases SdgA and SdgB mediate immunogenicity and protection of virulence-associated cell wall proteins
- PMID: 24130480
- PMCID: PMC3794999
- DOI: 10.1371/journal.ppat.1003653
Novel staphylococcal glycosyltransferases SdgA and SdgB mediate immunogenicity and protection of virulence-associated cell wall proteins
Abstract
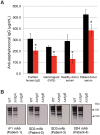
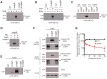
Infection of host tissues by Staphylococcus aureus and S. epidermidis requires an unusual family of staphylococcal adhesive proteins that contain long stretches of serine-aspartate dipeptide-repeats (SDR). The prototype member of this family is clumping factor A (ClfA), a key virulence factor that mediates adhesion to host tissues by binding to extracellular matrix proteins such as fibrinogen. However, the biological siginificance of the SDR-domain and its implication for pathogenesis remain poorly understood. Here, we identified two novel bacterial glycosyltransferases, SdgA and SdgB, which modify all SDR-proteins in these two bacterial species. Genetic and biochemical data demonstrated that these two glycosyltransferases directly bind and covalently link N-acetylglucosamine (GlcNAc) moieties to the SDR-domain in a step-wise manner, with SdgB appending the sugar residues proximal to the target Ser-Asp repeats, followed by additional modification by SdgA. GlcNAc-modification of SDR-proteins by SdgB creates an immunodominant epitope for highly opsonic human antibodies, which represent up to 1% of total human IgG. Deletion of these glycosyltransferases renders SDR-proteins vulnerable to proteolysis by human neutrophil-derived cathepsin G. Thus, SdgA and SdgB glycosylate staphylococcal SDR-proteins, which protects them against host proteolytic activity, and yet generates major eptopes for the human anti-staphylococcal antibody response, which may represent an ongoing competition between host and pathogen.
Conflict of interest statement
WLWH, KKK, JHM, SML, QP, LRS, SR, JK, MX, IMS, TS, AS, YK, AO, CK, RV, EJB, MWT, and SM are employees of Genentech; MJK, TB, AQB, and HS are employees of AIMM Therapeutics; MS, KK and PSA are employees of Symphogen A/S; BAD has no competing interests. This does not alter our adherence to all PLoS Pathogens policies on sharing data and materials.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

