Rad52 sumoylation prevents the toxicity of unproductive Rad51 filaments independently of the anti-recombinase Srs2
- PMID: 24130504
- PMCID: PMC3794917
- DOI: 10.1371/journal.pgen.1003833
Rad52 sumoylation prevents the toxicity of unproductive Rad51 filaments independently of the anti-recombinase Srs2
Abstract
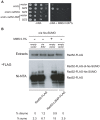
The budding yeast Srs2 is the archetype of helicases that regulate several aspects of homologous recombination (HR) to maintain genomic stability. Srs2 inhibits HR at replication forks and prevents high frequencies of crossing-over. Additionally, sensitivity to DNA damage and synthetic lethality with replication and recombination mutants are phenotypes that can only be attributed to another role of Srs2: the elimination of lethal intermediates formed by recombination proteins. To shed light on these intermediates, we searched for mutations that bypass the requirement of Srs2 in DNA repair without affecting HR. Remarkably, we isolated rad52-L264P, a novel allele of RAD52, a gene that encodes one of the most central recombination proteins in yeast. This mutation suppresses a broad spectrum of srs2Δ phenotypes in haploid cells, such as UV and γ-ray sensitivities as well as synthetic lethality with replication and recombination mutants, while it does not significantly affect Rad52 functions in HR and DNA repair. Extensive analysis of the genetic interactions between rad52-L264P and srs2Δ shows that rad52-L264P bypasses the requirement for Srs2 specifically for the prevention of toxic Rad51 filaments. Conversely, this Rad52 mutant cannot restore viability of srs2Δ cells that accumulate intertwined recombination intermediates which are normally processed by Srs2 post-synaptic functions. The avoidance of toxic Rad51 filaments by Rad52-L264P can be explained by a modification of its Rad51 filament mediator activity, as indicated by Chromatin immunoprecipitation and biochemical analysis. Remarkably, sensitivity to DNA damage of srs2Δ cells can also be overcome by stimulating Rad52 sumoylation through overexpression of the sumo-ligase SIZ2, or by replacing Rad52 by a Rad52-SUMO fusion protein. We propose that, like the rad52-L264P mutation, sumoylation modifies Rad52 activity thereby changing the properties of Rad51 filaments. This conclusion is strengthened by the finding that Rad52 is often associated with complete Rad51 filaments in vitro.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Rad52 Oligomeric N-Terminal Domain Stabilizes Rad51 Nucleoprotein Filaments and Contributes to Their Protection against Srs2.Cells. 2021 Jun 11;10(6):1467. doi: 10.3390/cells10061467. Cells. 2021. PMID: 34207997 Free PMC article.
-
Srs2 and Mus81-Mms4 Prevent Accumulation of Toxic Inter-Homolog Recombination Intermediates.PLoS Genet. 2016 Jul 7;12(7):e1006136. doi: 10.1371/journal.pgen.1006136. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27390022 Free PMC article.
-
Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase.J Biol Chem. 2009 Sep 4;284(36):24363-71. doi: 10.1074/jbc.M109.032953. Epub 2009 Jul 15. J Biol Chem. 2009. PMID: 19605344 Free PMC article.
-
Multifaceted role of the Saccharomyces cerevisiae Srs2 helicase in homologous recombination regulation.Biochem Soc Trans. 2005 Dec;33(Pt 6):1447-50. doi: 10.1042/BST0331447. Biochem Soc Trans. 2005. PMID: 16246143 Review.
-
Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair.FEMS Yeast Res. 2017 Mar 1;17(2):fow111. doi: 10.1093/femsyr/fow111. FEMS Yeast Res. 2017. PMID: 28011904 Free PMC article. Review.
Cited by
-
Caffeine inhibits gene conversion by displacing Rad51 from ssDNA.Nucleic Acids Res. 2015 Aug 18;43(14):6902-18. doi: 10.1093/nar/gkv525. Epub 2015 May 27. Nucleic Acids Res. 2015. PMID: 26019181 Free PMC article.
-
A Flp-SUMO hybrid recombinase reveals multi-layered copy number control of a selfish DNA element through post-translational modification.PLoS Genet. 2019 Jun 26;15(6):e1008193. doi: 10.1371/journal.pgen.1008193. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31242181 Free PMC article.
-
Single molecule microscopy reveals key physical features of repair foci in living cells.Elife. 2021 Feb 5;10:e60577. doi: 10.7554/eLife.60577. Elife. 2021. PMID: 33543712 Free PMC article.
-
Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae.Genetics. 2014 Nov;198(3):795-835. doi: 10.1534/genetics.114.166140. Genetics. 2014. PMID: 25381364 Free PMC article. Review.
-
Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52.Nat Commun. 2015 Jul 28;6:7834. doi: 10.1038/ncomms8834. Nat Commun. 2015. PMID: 26215801 Free PMC article.
References
-
- Cerbinskaite A, Mukhopadhyay A, Plummer ER, Curtin NJ, Edmondson RJ (2011) Defective homologous recombination in human cancers. Cancer Treat Rev 38: 89–100. - PubMed
-
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271. - PubMed
-
- San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

