Inhibition of the mitotic exit network in response to damaged telomeres
- PMID: 24130507
- PMCID: PMC3794921
- DOI: 10.1371/journal.pgen.1003859
Inhibition of the mitotic exit network in response to damaged telomeres
Abstract
When chromosomal DNA is damaged, progression through the cell cycle is halted to provide the cells with time to repair the genetic material before it is distributed between the mother and daughter cells. In Saccharomyces cerevisiae, this cell cycle arrest occurs at the G2/M transition. However, it is also necessary to restrain exit from mitosis by maintaining Bfa1-Bub2, the inhibitor of the Mitotic Exit Network (MEN), in an active state. While the role of Bfa1 and Bub2 in the inhibition of mitotic exit when the spindle is not properly aligned and the spindle position checkpoint is activated has been extensively studied, the mechanism by which these proteins prevent MEN function after DNA damage is still unclear. Here, we propose that the inhibition of the MEN is specifically required when telomeres are damaged but it is not necessary to face all types of chromosomal DNA damage, which is in agreement with previous data in mammals suggesting the existence of a putative telomere-specific DNA damage response that inhibits mitotic exit. Furthermore, we demonstrate that the mechanism of MEN inhibition when telomeres are damaged relies on the Rad53-dependent inhibition of Bfa1 phosphorylation by the Polo-like kinase Cdc5, establishing a new key role of this kinase in regulating cell cycle progression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
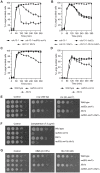
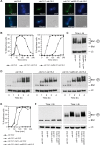
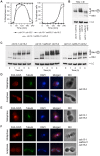
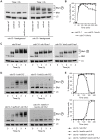
Similar articles
-
Asymmetry of the budding yeast Tem1 GTPase at spindle poles is required for spindle positioning but not for mitotic exit.PLoS Genet. 2015 Feb 6;11(2):e1004938. doi: 10.1371/journal.pgen.1004938. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25658911 Free PMC article.
-
Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints.Cell. 2001 Nov 30;107(5):655-65. doi: 10.1016/s0092-8674(01)00580-3. Cell. 2001. PMID: 11733064
-
Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints.Cell Cycle. 2002 Sep-Oct;1(5):351-5. Cell Cycle. 2002. PMID: 12461298
-
Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1.Small GTPases. 2015 Oct 2;6(4):196-201. doi: 10.1080/21541248.2015.1109023. Small GTPases. 2015. PMID: 26507466 Free PMC article. Review.
-
Regulation of Mitotic Exit by Cell Cycle Checkpoints: Lessons From Saccharomyces cerevisiae.Genes (Basel). 2020 Feb 12;11(2):195. doi: 10.3390/genes11020195. Genes (Basel). 2020. PMID: 32059558 Free PMC article. Review.
Cited by
-
Adaptation in replicative senescence: a risky business.Curr Genet. 2019 Jun;65(3):711-716. doi: 10.1007/s00294-019-00933-7. Epub 2019 Jan 12. Curr Genet. 2019. PMID: 30637477 Review.
-
Functions and regulation of the Polo-like kinase Cdc5 in the absence and presence of DNA damage.Curr Genet. 2018 Feb;64(1):87-96. doi: 10.1007/s00294-017-0727-2. Epub 2017 Aug 2. Curr Genet. 2018. PMID: 28770345 Free PMC article. Review.
-
Reduced kinase activity of polo kinase Cdc5 affects chromosome stability and DNA damage response in S. cerevisiae.Cell Cycle. 2016 Nov;15(21):2906-2919. doi: 10.1080/15384101.2016.1222338. Epub 2016 Aug 26. Cell Cycle. 2016. PMID: 27565373 Free PMC article.
-
Checkpoint adaptation: Keeping Cdc5 in the T-loop.Cell Cycle. 2016 Dec 16;15(24):3339-3340. doi: 10.1080/15384101.2016.1237769. Epub 2016 Sep 29. Cell Cycle. 2016. PMID: 27686022 Free PMC article. No abstract available.
-
The budding yeast Polo-like kinase localizes to distinct populations at centrosomes during mitosis.Mol Biol Cell. 2017 Apr 15;28(8):1011-1020. doi: 10.1091/mbc.E16-05-0324. Epub 2017 Feb 22. Mol Biol Cell. 2017. PMID: 28228549 Free PMC article.
References
-
- Harrison JC, Haber JE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235. - PubMed
-
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393. - PubMed
-
- Hu F, Wang Y, Liu D, Li Y, Qin J, et al. (2001) Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107: 655–665. - PubMed
-
- Stegmeier F, Amon A (2004) Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet 38: 203–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

