Identification of candidate B-lymphoma genes by cross-species gene expression profiling
- PMID: 24130802
- PMCID: PMC3793908
- DOI: 10.1371/journal.pone.0076889
Identification of candidate B-lymphoma genes by cross-species gene expression profiling
Abstract
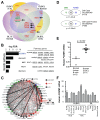
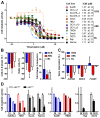
Comparative genome-wide expression profiling of malignant tumor counterparts across the human-mouse species barrier has a successful track record as a gene discovery tool in liver, breast, lung, prostate and other cancers, but has been largely neglected in studies on neoplasms of mature B-lymphocytes such as diffuse large B cell lymphoma (DLBCL) and Burkitt lymphoma (BL). We used global gene expression profiles of DLBCL-like tumors that arose spontaneously in Myc-transgenic C57BL/6 mice as a phylogenetically conserved filter for analyzing the human DLBCL transcriptome. The human and mouse lymphomas were found to have 60 concordantly deregulated genes in common, including 8 genes that Cox hazard regression analysis associated with overall survival in a published landmark dataset of DLBCL. Genetic network analysis of the 60 genes followed by biological validation studies indicate FOXM1 as a candidate DLBCL and BL gene, supporting a number of studies contending that FOXM1 is a therapeutic target in mature B cell tumors. Our findings demonstrate the value of the "mouse filter" for genomic studies of human B-lineage neoplasms for which a vast knowledge base already exists.
Conflict of interest statement
Figures



References
-
- Gaspar C, Cardoso J, Franken P, Molenaar L, Morreau H et al. (2008) Cross-species comparison of human and mouse intestinal polyps reveals conserved mechanisms in adenomatous polyposis coli (APC)-driven tumorigenesis. Am J Pathol 172: 1363-1380. doi:10.2353/ajpath.2008.070851. PubMed: 18403596. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

