Deep bisulfite sequencing of aberrantly methylated loci in a patient with multiple methylation defects
- PMID: 24130816
- PMCID: PMC3793946
- DOI: 10.1371/journal.pone.0076953
Deep bisulfite sequencing of aberrantly methylated loci in a patient with multiple methylation defects
Erratum in
- PLoS One. 2013;8(12). doi:10.1371/annotation/8a05c514-48b6-4dd9-b322-273b575cbd95
Abstract
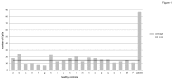
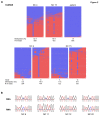
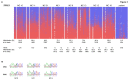
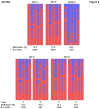
NLRP7 is a maternal effect gene as maternal mutations in this gene cause recurrent hydatidiform moles, spontaneous abortions and stillbirths, whereas live births are very rare. We have studied a patient with multiple anomalies born to a mother with a heterozygous NLRP7 mutation. By array-based CpG methylation analysis of blood DNA from the patient, his parents and 18 normal controls on Illumina Infinium HumanMethylation27 BeadChips we found that the patient had methylation changes (delta ß ≥ 0.3) at many imprinted loci as well as at 87 CpGs associated with 85 genes of unknown imprinting status. Using a pseudoproband (permutation) approach, we found methylation changes at only 7-24 CpGs (mean 15; standard deviation 4.84) in the controls. Thus, the number of abberantly methylated CpGs in the patient is more than 14 standard deviations higher. In order to identify novel imprinted genes among the 85 conspicuous genes in the patient, we selected 19 (mainly hypomethylated) genes for deep bisulfite amplicon sequencing on the ROCHE/454 Genome Sequencer in the patient and at least two additional controls. These controls had not been included in the array analysis and were heterozygous for a single nucleotide polymorphism at the test locus, so that allele-specific DNA methylation patterns could be determined. Apart from FAM50B, which we proved to be imprinted in blood, we did not observe allele-specific DNA methylation at the other 18 loci. We conclude that the patient does not only have methylation defects at imprinted loci but (at least in blood) also an excess of methylation changes at apparently non-imprinted loci.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

