Combined genetic and telemetry data reveal high rates of gene flow, migration, and long-distance dispersal potential in Arctic ringed seals (Pusa hispida)
- PMID: 24130843
- PMCID: PMC3794998
- DOI: 10.1371/journal.pone.0077125
Combined genetic and telemetry data reveal high rates of gene flow, migration, and long-distance dispersal potential in Arctic ringed seals (Pusa hispida)
Abstract
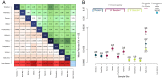
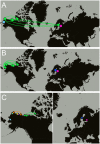
Ringed seals (Pusa hispida) are broadly distributed in seasonally ice covered seas, and their survival and reproductive success is intricately linked to sea ice and snow. Climatic warming is diminishing Arctic snow and sea ice and threatens to endanger ringed seals in the foreseeable future. We investigated the population structure and connectedness within and among three subspecies: Arctic (P. hispida hispida), Baltic (P. hispida botnica), and Lake Saimaa (P. hispida saimensis) ringed seals to assess their capacity to respond to rapid environmental changes. We consider (a) the geographical scale of migration, (b) use of sea ice, and (c) the amount of gene flow between subspecies. Seasonal movements and use of sea ice were determined for 27 seals tracked via satellite telemetry. Additionally, population genetic analyses were conducted using 354 seals representative of each subspecies and 11 breeding sites. Genetic analyses included sequences from two mitochondrial regions and genotypes of 9 microsatellite loci. We found that ringed seals disperse on a pan-Arctic scale and both males and females may migrate long distances during the summer months when sea ice extent is minimal. Gene flow among Arctic breeding sites and between the Arctic and the Baltic Sea subspecies was high; these two subspecies are interconnected as are breeding sites within the Arctic subspecies.
Conflict of interest statement
Figures





References
-
- Moore SE, Huntington HP (2008) Arctic marine mammals and climate change: impacts and resilience. Ecological Applications 18: s157–165. - PubMed
-
- Post E, Forchhammer MC, Bret-Harte MS, Callaghan TV, Christensen TR, et al. (2009) Ecological dynamics across the Arctic associated with recent climate change. Science 325: 1355–1358. - PubMed
-
- Monnett C, Gleason JS (2006) Observations of mortality associated with extended open-water swimming by polar bears in the Alaskan Beaufort Sea. Polar Biology 29: 681–687.
-
- United States Fish and Wildlife Service (2008) Endangered and threatened wildlife and plants; determination of threatened status for the polar bear (Ursus maritimus) throughout its range.
-
- Ferguson S, Stirling I, McLoughlin P (2005) Climate Change and ringed seal (Phoca hispida) recruitment in Western Hudson Bay. Marine Mammal Science 21: 121–135.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

