HIV's Nef interacts with β-catenin of the Wnt signaling pathway in HEK293 cells
- PMID: 24130899
- PMCID: PMC3795062
- DOI: 10.1371/journal.pone.0077865
HIV's Nef interacts with β-catenin of the Wnt signaling pathway in HEK293 cells
Abstract
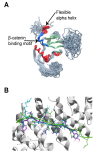
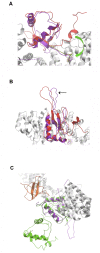
The Wnt signaling pathway is implicated in major physiologic cellular functions, such as proliferation, migration, cell fate specification, maintenance of pluripotency and induction of tumorigenicity. Proliferation and migration are important responses of T-cells, which are major cellular targets of HIV infection. Using an informatics screen, we identified a previously unsuspected interaction between HIV's Nef protein and β-catenin, a key component of the Wnt pathway. A segment in Nef contains identical amino acids at key positions and structurally mimics the β-catenin binding sites on endogenous β-catenin ligands. The interaction between Nef and β-catenin was confirmed in vitro and in a co-immunoprecipitation from HEK293 cells. Moreover, the introduction of Nef into HEK293 cells specifically inhibited a Wnt pathway reporter.
Conflict of interest statement
Figures



Similar articles
-
D186/D190 is an allele-dependent determinant of HIV-1 Nef function.Virology. 2016 Nov;498:44-56. doi: 10.1016/j.virol.2016.08.012. Epub 2016 Aug 24. Virology. 2016. PMID: 27560372
-
Secretion modification region-derived peptide disrupts HIV-1 Nef's interaction with mortalin and blocks virus and Nef exosome release.J Virol. 2012 Jan;86(1):406-19. doi: 10.1128/JVI.05720-11. Epub 2011 Oct 19. J Virol. 2012. PMID: 22013042 Free PMC article.
-
The ABCA1 domain responsible for interaction with HIV-1 Nef is conformational and not linear.Biochem Biophys Res Commun. 2014 Jan 31;444(1):19-23. doi: 10.1016/j.bbrc.2013.12.141. Epub 2014 Jan 7. Biochem Biophys Res Commun. 2014. PMID: 24406162 Free PMC article.
-
How HIV Nef Proteins Hijack Membrane Traffic To Promote Infection.J Virol. 2019 Nov 26;93(24):e01322-19. doi: 10.1128/JVI.01322-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31578291 Free PMC article. Review.
-
Physiological relevance of ACOT8-Nef interaction in HIV infection.Rev Med Virol. 2019 Sep;29(5):e2057. doi: 10.1002/rmv.2057. Epub 2019 Jun 9. Rev Med Virol. 2019. PMID: 31179598 Review.
Cited by
-
Resistance to the Tat Inhibitor Didehydro-Cortistatin A Is Mediated by Heightened Basal HIV-1 Transcription.mBio. 2019 Jul 2;10(4):e01750-18. doi: 10.1128/mBio.01750-18. mBio. 2019. PMID: 31266880 Free PMC article.
-
Regulation of influenza virus replication by Wnt/β-catenin signaling.PLoS One. 2018 Jan 11;13(1):e0191010. doi: 10.1371/journal.pone.0191010. eCollection 2018. PLoS One. 2018. PMID: 29324866 Free PMC article.
-
People living with HIV have low trabecular bone mineral density, high bone marrow adiposity, and poor trabecular bone microarchitecture at the proximal femur.Osteoporos Int. 2022 Aug;33(8):1739-1753. doi: 10.1007/s00198-022-06405-y. Epub 2022 Apr 27. Osteoporos Int. 2022. PMID: 35478045 Free PMC article.
-
Beta-catenin inhibits bovine parainfluenza virus type 3 replication via innate immunity pathway.BMC Vet Res. 2020 Mar 3;16(1):72. doi: 10.1186/s12917-020-02291-w. BMC Vet Res. 2020. PMID: 32127006 Free PMC article.
-
Loss of In Vivo Replication Fitness of HIV-1 Variants Resistant to the Tat Inhibitor, dCA.Viruses. 2023 Apr 12;15(4):950. doi: 10.3390/v15040950. Viruses. 2023. PMID: 37112931 Free PMC article.
References
-
- Churchill MJ, Rhodes DI, Learmont JC, Sullivan JS, Wesselingh SL et al. (2006) Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J Virol 80: 1047-1052. doi:10.1128/JVI.80.2.1047-1052.2006. PubMed: 16379007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

