Functionally and phenotypically distinct subpopulations of marrow stromal cells are fibroblast in origin and induce different fates in peripheral blood monocytes
- PMID: 24131213
- PMCID: PMC3967370
- DOI: 10.1089/scd.2013.0300
Functionally and phenotypically distinct subpopulations of marrow stromal cells are fibroblast in origin and induce different fates in peripheral blood monocytes
Abstract
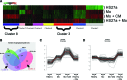
Marrow stromal cells constitute a heterogeneous population of cells, typically isolated after expansion in culture. In vivo, stromal cells often exist in close proximity or in direct contact with monocyte-derived macrophages, yet their interaction with monocytes is largely unexplored. In this report, isolated CD146(+) and CD146(-) stromal cells, as well as immortalized cell lines representative of each (designated HS27a and HS5, respectively), were shown by global DNase I hypersensitive site mapping and principal coordinate analysis to have a lineage association with marrow fibroblasts. Gene expression profiles generated for the CD146(+) and CD146(-) cell lines indicate significant differences in their respective transcriptomes, which translates into differences in secreted factors. Consequently, the conditioned media (CM) from these two populations induce different fates in peripheral blood monocytes. Monocytes incubated in CD146(+) CM acquire a tissue macrophage phenotype, whereas monocytes incubated in CM from CD146(-) cells express markers associated with pre-dendritic cells. Importantly, when CD14(+) monocytes are cultured in contact with the CD146(+) cells, the combined cell populations, assayed as a unit, show increased levels of transcripts associated with organismal development and hematopoietic regulation. In contrast, the gene expression profile from cocultures of monocytes and CD146(-) cells does not differ from that obtained when monocytes are cultured with CD146(-) CM. These in vitro results show that the CD146(+) marrow stromal cells together with monocytes increase the expression of genes relevant to hematopoietic regulation. In vivo relevance of these data is suggested by immunohistochemistry of marrow biopsies showing juxtaposed CD146(+) cells and CD68(+) cells associated with these upregulated proteins.
Figures




References
-
- Dexter TM. (1982). Stromal cell associated haemopoiesis. J Cell Physiol 1:87–94 - PubMed
-
- Allen TD. and Dexter TM. (1984). The essential cells of the hemopoietic microenvironment. Exp Hematol 12:517–521 - PubMed
-
- Dexter TM. and Moore MAS. (1977). In vitro duplication and “cure”. of haemopoietic defects in genetically anaemic mice. Nature 269:412–414 - PubMed
-
- Raaijmakers MH. and Scadden DT. (2008). Evolving concepts on the microenvironmental niche for hematopoietic stem cells. Curr Opin Hematol 15:301–306 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

