Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective
- PMID: 24133488
- PMCID: PMC3796261
- DOI: 10.3389/fmicb.2013.00303
Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective
Abstract
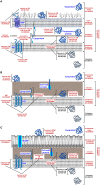
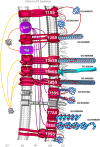
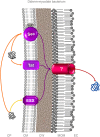
Bacterial colonization of biotic or abiotic surfaces results from two quite distinct physiological processes, namely bacterial adhesion and biofilm formation. Broadly speaking, a biofilm is defined as the sessile development of microbial cells. Biofilm formation arises following bacterial adhesion but not all single bacterial cells adhering reversibly or irreversibly engage inexorably into a sessile mode of growth. Among molecular determinants promoting bacterial colonization, surface proteins are the most functionally diverse active components. To be present on the bacterial cell surface, though, a protein must be secreted in the first place. Considering the close association of secreted proteins with their cognate secretion systems, the secretome (which refers both to the secretion systems and their protein substrates) is a key concept to apprehend the protein secretion and related physiological functions. The protein secretion systems are here considered in light of the differences in the cell-envelope architecture between diderm-LPS (archetypal Gram-negative), monoderm (archetypal Gram-positive) and diderm-mycolate (archetypal acid-fast) bacteria. Besides, their cognate secreted proteins engaged in the bacterial colonization process are regarded from single protein to supramolecular protein structure as well as the non-classical protein secretion. This state-of-the-art on the complement of the secretome (the secretion systems and their cognate effectors) involved in the surface colonization process in diderm-LPS and monoderm bacteria paves the way for future research directions in the field.
Keywords: MSCRAMM; adhesin; aggregation; cell surface; pili/fimbriae/curli; protein secretion system; secreted protein; secretome.
Figures

References
-
- An Y. H., Friedman R. J. (1998). Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J. Biomed. Mater. Res. 43, 338–348 - PubMed
-
- Antelmann H., Van Dijl J. M., Bron S., Hecker M. (2006). Proteomic survey through secretome of Bacillus subtilis. Methods Biochem. Anal. 49, 179–208 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

