Ensemble of gene signatures identifies novel biomarkers in colorectal cancer activated through PPARγ and TNFα signaling
- PMID: 24133572
- PMCID: PMC3795784
- DOI: 10.1371/journal.pone.0072638
Ensemble of gene signatures identifies novel biomarkers in colorectal cancer activated through PPARγ and TNFα signaling
Abstract
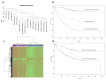
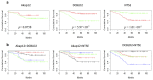
We describe a novel bioinformatic and translational pathology approach, gene Signature Finder Algorithm (gSFA) to identify biomarkers associated with Colorectal Cancer (CRC) survival. Here a robust set of CRC markers is selected by an ensemble method. By using a dataset of 232 gene expression profiles, gSFA discovers 16 highly significant small gene signatures. Analysis of dichotomies generated by the signatures results in a set of 133 samples stably classified in good prognosis group and 56 samples in poor prognosis group, whereas 43 remain unreliably classified. AKAP12, DCBLD2, NT5E and SPON1 are particularly represented in the signatures and selected for validation in vivo on two independent patients cohorts comprising 140 tumor tissues and 60 matched normal tissues. Their expression and regulatory programs are investigated in vitro. We show that the coupled expression of NT5E and DCBLD2 robustly stratifies our patients in two groups (one of which with 100% survival at five years). We show that NT5E is a target of the TNF-α signaling in vitro; the tumor suppressor PPARγ acts as a novel NT5E antagonist that positively and concomitantly regulates DCBLD2 in a cancer cell context-dependent manner.
Conflict of interest statement
Figures



References
-
- Cancer Genome Atlas Network (2012). Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 487: 330–337
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

