Paralogue annotation identifies novel pathogenic variants in patients with Brugada syndrome and catecholaminergic polymorphic ventricular tachycardia
- PMID: 24136861
- PMCID: PMC3888601
- DOI: 10.1136/jmedgenet-2013-101917
Paralogue annotation identifies novel pathogenic variants in patients with Brugada syndrome and catecholaminergic polymorphic ventricular tachycardia
Abstract
Background: Distinguishing genetic variants that cause disease from variants that are rare but benign is one of the principal challenges in contemporary clinical genetics, particularly as variants are identified at a pace exceeding the capacity of researchers to characterise them functionally.
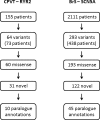
Methods: We previously developed a novel method, called paralogue annotation, which accurately and specifically identifies disease-causing missense variants by transferring disease-causing annotations across families of related proteins. Here we refine our approach, and apply it to novel variants found in 2266 patients across two large cohorts with inherited sudden death syndromes, namely catecholaminergic polymorphic ventricular tachycardia (CPVT) or Brugada syndrome (BrS).
Results: Over one third of the novel non-synonymous variants found in these studies, which would otherwise be reported in a clinical diagnostics setting as 'variants of unknown significance', are categorised by our method as likely disease causing (positive predictive value 98.7%). This identified more than 500 new disease loci for BrS and CPVT.
Conclusions: Our methodology is widely transferable across all human disease genes, with an estimated 150 000 potentially informative annotations in more than 1800 genes. We have developed a web resource that allows researchers and clinicians to annotate variants found in individuals with inherited arrhythmias, comprising a referenced compendium of known missense variants in these genes together with a user-friendly implementation of our approach. This tool will facilitate the interpretation of many novel variants that might otherwise remain unclassified.
Keywords: Cardiovascular Medicine; Clinical Genetics; Congenital Heart Disease; Genetics.
Figures


References
-
- Alings M, Wilde A. Brugada syndrome: clinical data and suggested pathophysiological mechanism. Circulation 1999;99:666–73 - PubMed
-
- Priori SG, Napolitano C, Gasparini M, Pappone C, Della Bella P, Giordano U, Bloise R, Giustetto C, De Nardis R, Grillo M, Ronchetti E, Faggiano G, Nastoli J. Natural history of Brugada syndrome: insights for risk stratification and management. Circulation 2002;105:1342–7 - PubMed
-
- Priori SG, Napolitano C, Memmi M, Colombi B, Drago F, Gasparini M, DeSimone L, Coltorti F, Bloise R, Keegan R, Cruz Filho FE, Vignati G, Benatar A, DeLogu A. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation 2002;106:69–74 - PubMed
-
- Medeiros-Domingo A, Bhuiyan ZA, Tester DJ, Hofman N, Bikker H, van Tintelen JP, Mannens MM, Wilde AA, Ackerman MJ. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: a comprehensive open reading frame mutational analysis. J Am Coll Cardiol 2009;54:2065–74 - PMC - PubMed
-
- Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J, Brugada P, Fressart V, Guerchicoff A, Harris-Kerr C, Kamakura S, Kyndt F, Koopmann TT, Miyamoto Y, Pfeiffer R, Pollevick GD, Probst V, Zumhagen S, Vatta M, Towbin JA, Shimizu W, Schulze-Bahr E, Antzelevitch C, Salisbury BA, Guicheney P, Wilde AA, Brugada R, Schott JJ, Ackerman MJ. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 2009;7:33–46 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
