Genomically recoded organisms expand biological functions
- PMID: 24136966
- PMCID: PMC4924538
- DOI: 10.1126/science.1241459
Genomically recoded organisms expand biological functions
Abstract
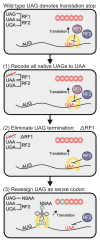
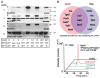
We describe the construction and characterization of a genomically recoded organism (GRO). We replaced all known UAG stop codons in Escherichia coli MG1655 with synonymous UAA codons, which permitted the deletion of release factor 1 and reassignment of UAG translation function. This GRO exhibited improved properties for incorporation of nonstandard amino acids that expand the chemical diversity of proteins in vivo. The GRO also exhibited increased resistance to T7 bacteriophage, demonstrating that new genetic codes could enable increased viral resistance.
Figures
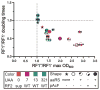
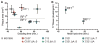
 ) ΔprfA::tolC (ΔA::T); and (×) a clean deletion of prfA (ΔA). (A) RF1 status affects plaque area (Kruskal-Wallis one-way analysis of variance, P < 0.001), but strain doubling time does not (Pearson correlation, P = 0.49). Plaque areas (mm2) were calculated with ImageJ, and means ± 95% confidence intervals are reported (n > 12 for each strain). In the absence of RF1, all strains except C0.B*.ΔA::S yielded significantly smaller plaques, indicating that the RF2 variant (16) can terminate UAG adequately to maintain T7 fitness. A statistical summary can be found in table S14. (B) T7 fitness (doublings/hour) (22) is impaired (P = 0.002) and mean lysis time (min) is increased (P < 0.0001) in C321.ΔA compared to C321. Significance was assessed for each metric by using an unpaired t test with Welch’s correction.
) ΔprfA::tolC (ΔA::T); and (×) a clean deletion of prfA (ΔA). (A) RF1 status affects plaque area (Kruskal-Wallis one-way analysis of variance, P < 0.001), but strain doubling time does not (Pearson correlation, P = 0.49). Plaque areas (mm2) were calculated with ImageJ, and means ± 95% confidence intervals are reported (n > 12 for each strain). In the absence of RF1, all strains except C0.B*.ΔA::S yielded significantly smaller plaques, indicating that the RF2 variant (16) can terminate UAG adequately to maintain T7 fitness. A statistical summary can be found in table S14. (B) T7 fitness (doublings/hour) (22) is impaired (P = 0.002) and mean lysis time (min) is increased (P < 0.0001) in C321.ΔA compared to C321. Significance was assessed for each metric by using an unpaired t test with Welch’s correction.References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

