Identification, nomenclature, and evolutionary relationships of mitogen-activated protein kinase (MAPK) genes in soybean
- PMID: 24137047
- PMCID: PMC3785387
- DOI: 10.4137/EBO.S12526
Identification, nomenclature, and evolutionary relationships of mitogen-activated protein kinase (MAPK) genes in soybean
Abstract
Mitogen-activated protein kinase (MAPK) genes in eukaryotes regulate various developmental and physiological processes including those associated with biotic and abiotic stresses. Although MAPKs in some plant species including Arabidopsis have been identified, they are yet to be identified in soybean. Major objectives of this study were to identify GmMAPKs, assess their evolutionary relationships, and analyze their functional divergence. We identified a total of 38 MAPKs, eleven MAPKKs, and 150 MAPKKKs in soybean. Within the GmMAPK family, we also identified a new clade of six genes: four genes with TEY and two genes with TQY motifs requiring further investigation into possible legume-specific functions. The results indicated the expansion of the GmMAPK families attributable to the ancestral polyploidy events followed by chromosomal rearrangements. The GmMAPK and GmMAPKKK families were substantially larger than those in other plant species. The duplicated GmMAPK members presented complex evolutionary relationships and functional divergence when compared to their counterparts in Arabidopsis. We also highlighted existing nomenclatural issues, stressing the need for nomenclatural consistency. GmMAPK identification is vital to soybean crop improvement, and novel insights into the evolutionary relationships will enhance our understanding about plant genome evolution.
Keywords: MAPK family; gene evolution; homology; nomenclature; signal transduction; soybean genomics.
Figures
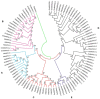



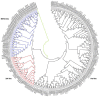









References
-
- Rohila JS, Yang Y. Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. J Integr Plant Biol. 2007;49(6):751–9.
-
- Jonak C, Okrész L, Bögre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002;5(5):415–24. - PubMed
-
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–49. - PubMed
-
- Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–13. - PubMed
-
- Huang GT, Ma SL, Bai LP, et al. Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep. 2012;39(2):969–87. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

