Intestinal epithelium and autophagy: partners in gut homeostasis
- PMID: 24137160
- PMCID: PMC3786390
- DOI: 10.3389/fimmu.2013.00301
Intestinal epithelium and autophagy: partners in gut homeostasis
Abstract
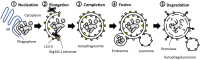
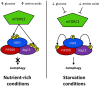
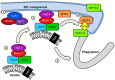
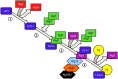
One of the most significant challenges of cell biology is to understand how each type of cell copes with its specific workload without suffering damage. Among the most intriguing questions concerns intestinal epithelial cells in mammals; these cells act as a barrier between the internally protected region and the external environment that is exposed constantly to food and microbes. A major process involved in the processing of microbes is autophagy. In the intestine, through multiple, complex signaling pathways, autophagy including macroautophagy and xenophagy is pivotal in mounting appropriate intestinal immune responses and anti-microbial protection. Dysfunctional autophagy mechanism leads to chronic intestinal inflammation, such as inflammatory bowel disease (IBD). Studies involving a number of in vitro and in vivo mouse models in addition to human clinical studies have revealed a detailed role for autophagy in the generation of chronic intestinal inflammation. A number of genome-wide association studies identified roles for numerous autophagy genes in IBD, especially in Crohn's disease. In this review, we will explore in detail the latest research linking autophagy to intestinal homeostasis and how alterations in autophagy pathways lead to intestinal inflammation.
Keywords: ATG16L1; IBD; IRGM; autophagy; intestinal epithelium.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

