Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development
- PMID: 24137171
- PMCID: PMC3797442
- DOI: 10.3389/fpls.2013.00406
Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development
Abstract
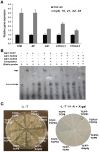
TCP family of plant-specific transcription factors regulates plant form through control of cell proliferation and differentiation. This gene family is comprised of two groups, class I and class II. While the role of class II TCP genes in plant development is well known, data about the function of some class I TCP genes is lacking. We studied a group of phylogenetically related class I TCP genes: AtTCP7, AtTCP8, AtTCP22, and AtTCP23. The similar expression pattern in young growing leaves found for this group suggests similarity in gene function. Gene redundancy is characteristic in this group, as also seen in the class II TCP genes. We generated a pentuple mutant tcp8 tcp15 tcp21 tcp22 tcp23 and show that loss of function of these genes results in changes in leaf developmental traits. We also determined that these factors are able to mutually interact in a yeast two-hybrid assay and regulate the expression of KNOX1 genes. To circumvent the issue of genetic redundancy, dominant negative forms with SRDX repressor domain were used. Analysis of transgenic plants expressing AtTCP7-SRDX and AtTCP23-SRDX indicate a role of these factors in the control of cell proliferation.
Keywords: Arabidopsis; SRDX construct; TCP; leaf development; transcription factor.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

