A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity
- PMID: 24137537
- PMCID: PMC3793227
- DOI: 10.7554/eLife.00994
A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity
Abstract
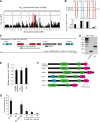
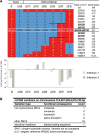
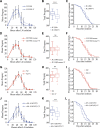
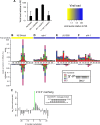
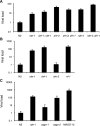
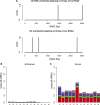
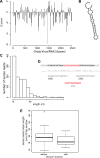
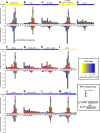
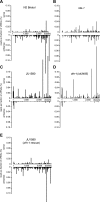
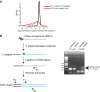
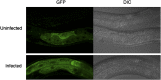
RNA interference defends against viral infection in plant and animal cells. The nematode Caenorhabditis elegans and its natural pathogen, the positive-strand RNA virus Orsay, have recently emerged as a new animal model of host-virus interaction. Using a genome-wide association study in C. elegans wild populations and quantitative trait locus mapping, we identify a 159 base-pair deletion in the conserved drh-1 gene (encoding a RIG-I-like helicase) as a major determinant of viral sensitivity. We show that DRH-1 is required for the initiation of an antiviral RNAi pathway and the generation of virus-derived siRNAs (viRNAs). In mammals, RIG-I-domain containing proteins trigger an interferon-based innate immunity pathway in response to RNA virus infection. Our work in C. elegans demonstrates that the RIG-I domain has an ancient role in viral recognition. We propose that RIG-I acts as modular viral recognition factor that couples viral recognition to different effector pathways including RNAi and interferon responses. DOI:http://dx.doi.org/10.7554/eLife.00994.001.
Keywords: C. elegans; RNA interference; immunity; virus infection.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

