A novel type of N-acetylglutamate synthase is involved in the first step of arginine biosynthesis in Corynebacterium glutamicum
- PMID: 24138314
- PMCID: PMC3827942
- DOI: 10.1186/1471-2164-14-713
A novel type of N-acetylglutamate synthase is involved in the first step of arginine biosynthesis in Corynebacterium glutamicum
Abstract
Background: Arginine biosynthesis in Corynebacterium glutamicum consists of eight enzymatic steps, starting with acetylation of glutamate, catalysed by N-acetylglutamate synthase (NAGS). There are different kinds of known NAGSs, for example, "classical" ArgA, bifunctional ArgJ, ArgO, and S-NAGS. However, since C. glutamicum possesses a monofunctional ArgJ, which catalyses only the fifth step of the arginine biosynthesis pathway, glutamate must be acetylated by an as of yet unknown NAGS gene.
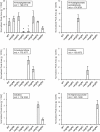
Results: Arginine biosynthesis was investigated by metabolome profiling using defined gene deletion mutants that were expected to accumulate corresponding intracellular metabolites. HPLC-ESI-qTOF analyses gave detailed insights into arginine metabolism by detecting six out of seven intermediates of arginine biosynthesis. Accumulation of N-acetylglutamate in all mutants was a further confirmation of the unknown NAGS activity. To elucidate the identity of this gene, a genomic library of C. glutamicum was created and used to complement an Escherichia coli ΔargA mutant. The plasmid identified, which allowed functional complementation, contained part of gene cg3035, which contains an acetyltransferase domain in its amino acid sequence. Deletion of cg3035 in the C. glutamicum genome led to a partial auxotrophy for arginine. Heterologous overexpression of the entire cg3035 gene verified its ability to complement the E. coli ΔargA mutant in vivo and homologous overexpression led to a significantly higher intracellular N-acetylglutamate pool. Enzyme assays confirmed the N-acetylglutamate synthase activity of Cg3035 in vitro. However, the amino acid sequence of Cg3035 revealed no similarities to members of known NAGS gene families.
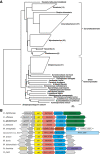
Conclusions: The N-acetylglutamate synthase Cg3035 is able to catalyse the first step of arginine biosynthesis in C. glutamicum. It represents a novel class of NAGS genes apparently present only in bacteria of the suborder Corynebacterineae, comprising amongst others the genera Corynebacterium, Mycobacterium, and Nocardia. Therefore, the name C-NAGS (Corynebacterineae-type NAGS) is proposed for this new family.
Figures



Similar articles
-
Identification of a suppressor gene for the arginine-auxotrophic argJ mutation in Corynebacterium glutamicum.J Ind Microbiol Biotechnol. 2010 Nov;37(11):1131-6. doi: 10.1007/s10295-010-0760-3. Epub 2010 Jun 12. J Ind Microbiol Biotechnol. 2010. PMID: 20544254
-
Bioinformatic analysis of an unusual gene-enzyme relationship in the arginine biosynthetic pathway among marine gamma proteobacteria: implications concerning the formation of N-acetylated intermediates in prokaryotes.BMC Genomics. 2006 Jan 12;7:4. doi: 10.1186/1471-2164-7-4. BMC Genomics. 2006. PMID: 16409639 Free PMC article.
-
A novel bifunctional N-acetylglutamate synthase-kinase from Xanthomonas campestris that is closely related to mammalian N-acetylglutamate synthase.BMC Biochem. 2007 Apr 10;8:4. doi: 10.1186/1471-2091-8-4. BMC Biochem. 2007. PMID: 17425781 Free PMC article.
-
The N-Acetylglutamate Synthase Family: Structures, Function and Mechanisms.Int J Mol Sci. 2015 Jun 9;16(6):13004-22. doi: 10.3390/ijms160613004. Int J Mol Sci. 2015. PMID: 26068232 Free PMC article. Review.
-
Surprising arginine biosynthesis: a reappraisal of the enzymology and evolution of the pathway in microorganisms.Microbiol Mol Biol Rev. 2007 Mar;71(1):36-47. doi: 10.1128/MMBR.00032-06. Microbiol Mol Biol Rev. 2007. PMID: 17347518 Free PMC article. Review.
Cited by
-
ALLocator: an interactive web platform for the analysis of metabolomic LC-ESI-MS datasets, enabling semi-automated, user-revised compound annotation and mass isotopomer ratio analysis.PLoS One. 2014 Nov 26;9(11):e113909. doi: 10.1371/journal.pone.0113909. eCollection 2014. PLoS One. 2014. PMID: 25426929 Free PMC article.
-
A novel strategy for L-arginine production in engineered Escherichia coli.Microb Cell Fact. 2023 Jul 26;22(1):138. doi: 10.1186/s12934-023-02145-8. Microb Cell Fact. 2023. PMID: 37495979 Free PMC article.
-
Improvement of L-citrulline production in Corynebacterium glutamicum by ornithine acetyltransferase.J Ind Microbiol Biotechnol. 2015 Feb;42(2):307-13. doi: 10.1007/s10295-014-1561-x. Epub 2014 Dec 10. J Ind Microbiol Biotechnol. 2015. PMID: 25492493
-
Metabolic engineering for efficient supply of acetyl-CoA from different carbon sources in Escherichia coli.Microb Cell Fact. 2019 Aug 6;18(1):130. doi: 10.1186/s12934-019-1177-y. Microb Cell Fact. 2019. PMID: 31387584 Free PMC article.
-
Transcriptomic investigations of polymyxins and colistin/sulbactam combination against carbapenem-resistant Acinetobacter baumannii.Comput Struct Biotechnol J. 2024 May 31;23:2595-2605. doi: 10.1016/j.csbj.2024.05.043. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39006922 Free PMC article.
References
-
- Glansdorff N, Ying X. In: Amino acid biosynthesis: pathways, regulation and metabolic engineering. Wendisch VF, editor. Wiesbaden: Springer; 2007. Microbial arginine biosynthesis: pathway, regulation and industrial production; pp. 219–257. [Steinbüchel A (Series editor): Microbiology Monographs, vol 5.]
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

