Integrative genomics identifies candidate microRNAs for pathogenesis of experimental biliary atresia
- PMID: 24138927
- PMCID: PMC3819657
- DOI: 10.1186/1752-0509-7-104
Integrative genomics identifies candidate microRNAs for pathogenesis of experimental biliary atresia
Abstract
Background: Biliary atresia is a fibroinflammatory obstruction of extrahepatic bile duct that leads to end-stage liver disease in children. Despite advances in understanding the pathogenesis of biliary atresia, very little is known about the role of microRNAs (miRNAs) in onset and progression of the disease. In this study, we aimed to investigate the entire biliary transcriptome to identify miRNAs with potential role in the pathogenesis of bile duct obstruction.
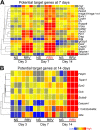
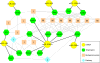
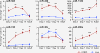
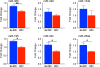
Results: By profiling the expression levels of miRNA in extrahepatic bile ducts and gallbladder (EHBDs) from a murine model of biliary atresia, we identified 14 miRNAs whose expression was suppressed at the times of duct obstruction and atresia (≥2 fold suppression, P < 0.05, FDR 5%). Next, we obtained 2,216 putative target genes of the 14 miRNAs using in silico target prediction algorithms. By integrating this result with a genome-wide gene expression analysis of the same tissue (≥2 fold increase, P < 0.05, FDR 5%), we identified 26 potential target genes with coordinate expression by the 14 miRNAs. Functional analysis of these target genes revealed a significant relevance of miR-30b/c, -133a/b, -195, -200a, -320 and -365 based on increases in expression of at least 3 target genes in the same tissue and 1st-to-3rd tier links with genes and gene-groups regulating organogenesis and immune response. These miRNAs showed higher expression in EHBDs above livers, a unique expression in cholangiocytes and the subepithelial compartment, and were downregulated in a cholangiocyte cell line after RRV infection.
Conclusions: Integrative genomics reveals functional relevance of miR-30b/c, -133a/b, -195, -200a, -320 and -365. The coordinate expression of miRNAs and target genes in a temporal-spatial fashion suggests a regulatory role of these miRNAs in pathogenesis of experimental biliary atresia.
Figures


References
-
- Carvalho E, Liu C, Shivakumar P, Sabla G, Aronow B, Bezerra JA. Analysis of the biliary transcriptome in experimental biliary atresia. Gastroenterology. 2005;129:713–717. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

