Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta
- PMID: 24139741
- PMCID: PMC3909816
- DOI: 10.1016/j.cub.2013.08.061
Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta
Abstract
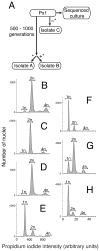
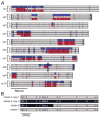
Nearly all animals reproduce sexually through the production and fusion of sperm and egg cells, yet little is known about the ancestry of animal sexual reproduction. Moreover, the sexual cycle of the closest living relatives of animals, the choanoflagellates, remains completely unknown. The choanoflagellate Monosiga brevicollis possesses a "meiotic toolkit" of genes, but the lack of polymorphisms detected during genome sequencing precluded inferences about its ploidy or sexual cycle. Here, we report that a related choanoflagellate, Salpingoeca rosetta, has a sexual life cycle and transitions between haploid and diploid states. Haploid cultures of S. rosetta became diploid in response to nutrient limitation. This ploidy shift coincided with anisogamous mating, during which small flagellated cells fused with larger flagellated cells. Distributions of polymorphisms in laboratory strains of S. rosetta provided independent evidence of historical recombination and mating. The ability of S. rosetta to produce morphologically differentiated gametes and to engage in sexual reproduction has implications for both reconstructing the evolution of sex in the progenitors of animals and establishing classical genetics in choanoflagellates.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures

Comment in
-
Evolution of sex: mating rituals of a pre-metazoan.Curr Biol. 2013 Nov 18;23(22):R1006-R1008. doi: 10.1016/j.cub.2013.10.009. Curr Biol. 2013. PMID: 24262825
References
-
- Carr M, Leadbeater BSC, Baldauf SL. Conserved Meiotic Genes Point to Sex in the Choanoflagellates. Journal of Eukaryotic Microbiology. 2010;57:56–62. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

