SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant
- PMID: 24140112
- PMCID: PMC3824129
- DOI: 10.1016/j.ajhg.2013.09.010
SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant
Erratum in
- Am J Hum Genet. 2013 Nov 7;93(5):994
Abstract
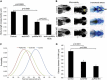
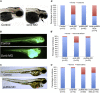
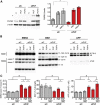
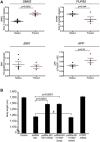
Copy-number variants (CNVs) represent a significant interpretative challenge, given that each CNV typically affects the dosage of multiple genes. Here we report on five individuals with coloboma, microcephaly, developmental delay, short stature, and craniofacial, cardiac, and renal defects who harbor overlapping microdeletions on 8q24.3. Fine mapping localized a commonly deleted 78 kb region that contains three genes: SCRIB, NRBP2, and PUF60. In vivo dissection of the CNV showed discrete contributions of the planar cell polarity effector SCRIB and the splicing factor PUF60 to the syndromic phenotype, and the combinatorial suppression of both genes exacerbated some, but not all, phenotypic components. Consistent with these findings, we identified an individual with microcephaly, short stature, intellectual disability, and heart defects with a de novo c.505C>T variant leading to a p.His169Tyr change in PUF60. Functional testing of this allele in vivo and in vitro showed that the mutation perturbs the relative dosage of two PUF60 isoforms and, subsequently, the splicing efficiency of downstream PUF60 targets. These data inform the functions of two genes not associated previously with human genetic disease and demonstrate how CNVs can exhibit complex genetic architecture, with the phenotype being the amalgam of both discrete dosage dysfunction of single transcripts and also of binary genetic interactions.
Copyright © 2013 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
-
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- McCarroll S.A., Altshuler D.M. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39(Suppl):S37–S42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

