Remodelling of a polypyrimidine tract-binding protein complex during apoptosis activates cellular IRESs
- PMID: 24141718
- PMCID: PMC3857619
- DOI: 10.1038/cdd.2013.135
Remodelling of a polypyrimidine tract-binding protein complex during apoptosis activates cellular IRESs
Abstract
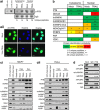
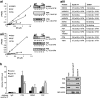
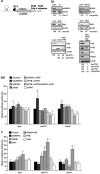
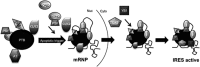
Post-transcriptional control of gene expression is mediated by the interaction of RNA-binding proteins with their cognate mRNAs that specifically regulate their stability, localization and translation. mRNA-binding proteins are multifunctional and it has been proposed therefore that a combinatorial RNA-binding protein code exists that allows specific protein sub-complexes to control cytoplasmic gene expression under a range of pathophysiological conditions. We show that polypyrimidine tract-binding protein (PTB) is central to one such complex that forms in apoptotic cells. Thus, during apoptosis initiated by TNF-related apoptosis inducing ligand there is a change in the repertoire of RNA-binding proteins with which PTB interacts. We show that altering the cellular levels of PTB and its binding partners, either singly or in combination, is sufficient to directly change the rates of apoptosis with increased expression of PTB, YBX1, PSF and NONO/p54(nrb) accelerating this process. Mechanistically, we show that these proteins post-transcriptionally regulate gene expression, and therefore apoptotic rates, by interacting with and stimulating the activity of RNA elements (internal ribosome entry segments) found in mRNAs that are translated during apoptosis. Taken together, our data show that PTB function is controlled by a set of co-recruited proteins and importantly provide further evidence that it is possible to dictate cell fate by modulating cytoplasmic gene expression pathways alone.
Figures



References
-
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. - PubMed
-
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, et al. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell. 2006;23:401–412. - PubMed
-
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 091595/WT_/Wellcome Trust/United Kingdom
- BB/F019017/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UP_A600_1024/MRC_/Medical Research Council/United Kingdom
- BB/F000065/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F011806/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

