Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing
- PMID: 24141982
- PMCID: PMC3883177
- DOI: 10.1093/infdis/jit546
Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing
Abstract
Background: The first comparison of a live RNA viral vaccine strain to its wild-type parental strain by deep sequencing is presented using as a model the yellow fever virus (YFV) live vaccine strain 17D-204 and its wild-type parental strain, Asibi.
Methods: The YFV 17D-204 vaccine genome was compared to that of the parental strain Asibi by massively parallel methods. Variability was compared on multiple scales of the viral genomes. A modeled exploration of small-frequency variants was performed to reconstruct plausible regions of mutational plasticity.
Results: Overt quasispecies diversity is a feature of the parental strain, whereas the live vaccine strain lacks diversity according to multiple independent measurements. A lack of attenuating mutations in the Asibi population relative to that of 17D-204 was observed, demonstrating that the vaccine strain was derived by discrete mutation of Asibi and not by selection of genomes in the wild-type population.
Conclusions: Relative quasispecies structure is a plausible correlate of attenuation for live viral vaccines. Analyses such as these of attenuated viruses improve our understanding of the molecular basis of vaccine attenuation and provide critical information on the stability of live vaccines and the risk of reversion to virulence.
Keywords: 17D, Viral Population; Deep Sequencing; Flavivirus; Parallel Sequencing; Quasispecies; Vaccine; Variants; Yellow Fever Virus.
Figures
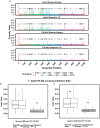
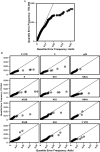
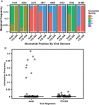
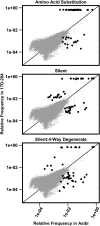
Comment in
-
Yellow fever vaccine attenuation revealed: loss of diversity.J Infect Dis. 2014 Feb 1;209(3):318-20. doi: 10.1093/infdis/jit551. Epub 2013 Oct 17. J Infect Dis. 2014. PMID: 24141980 No abstract available.
Similar articles
-
Attenuation of Live-Attenuated Yellow Fever 17D Vaccine Virus Is Localized to a High-Fidelity Replication Complex.mBio. 2019 Oct 22;10(5):e02294-19. doi: 10.1128/mBio.02294-19. mBio. 2019. PMID: 31641088 Free PMC article.
-
Genome Characterization of Yellow Fever Virus Wild-Type Strain Asibi, Parent to Live-Attenuated 17D Vaccine, from Three Different Sources.Viruses. 2021 Jul 16;13(7):1383. doi: 10.3390/v13071383. Viruses. 2021. PMID: 34372589 Free PMC article.
-
Vaccine and Wild-Type Strains of Yellow Fever Virus Engage Distinct Entry Mechanisms and Differentially Stimulate Antiviral Immune Responses.mBio. 2016 Feb 9;7(1):e01956-15. doi: 10.1128/mBio.01956-15. mBio. 2016. PMID: 26861019 Free PMC article.
-
The yellow fever 17D virus as a platform for new live attenuated vaccines.Hum Vaccin Immunother. 2014;10(5):1256-65. doi: 10.4161/hv.28117. Epub 2014 Feb 19. Hum Vaccin Immunother. 2014. PMID: 24553128 Free PMC article. Review.
-
The yellow fever 17D vaccine virus: molecular basis of viral attenuation and its use as an expression vector.Braz J Med Biol Res. 1997 Feb;30(2):157-68. doi: 10.1590/s0100-879x1997000200002. Braz J Med Biol Res. 1997. PMID: 9239300 Review.
Cited by
-
Engineering a fidelity-variant live-attenuated vaccine for chikungunya virus.NPJ Vaccines. 2020 Oct 14;5:97. doi: 10.1038/s41541-020-00241-z. eCollection 2020. NPJ Vaccines. 2020. PMID: 33083032 Free PMC article.
-
Guidelines for In Vitro Production and Quantification of Rift Valley Fever Virus.Methods Mol Biol. 2024;2824:91-104. doi: 10.1007/978-1-0716-3926-9_7. Methods Mol Biol. 2024. PMID: 39039408
-
Morphologic and Genetic Characterization of Ilheus Virus, a Potential Emergent Flavivirus in the Americas.Viruses. 2023 Jan 10;15(1):195. doi: 10.3390/v15010195. Viruses. 2023. PMID: 36680235 Free PMC article.
-
Analysis By Deep Sequencing of Discontinued Neurotropic Yellow Fever Vaccine Strains.Sci Rep. 2018 Sep 7;8(1):13408. doi: 10.1038/s41598-018-31085-2. Sci Rep. 2018. PMID: 30194325 Free PMC article.
-
Repurposing the yellow fever vaccine for intratumoral immunotherapy.EMBO Mol Med. 2020 Jan 9;12(1):e10375. doi: 10.15252/emmm.201910375. Epub 2019 Nov 19. EMBO Mol Med. 2020. PMID: 31746149 Free PMC article.
References
-
- Holland JJ, De La Torre JC, Steinhauer DA. RNA virus populations as quasispecies. Curr Top Microb Immunol. 1992;176:1–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

