The growth factor receptor ERBB2 regulates mitochondrial activity on a signaling time scale
- PMID: 24142693
- PMCID: PMC3853274
- DOI: 10.1074/jbc.M113.478271
The growth factor receptor ERBB2 regulates mitochondrial activity on a signaling time scale
Abstract
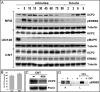
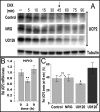
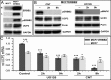
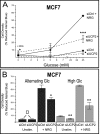
Overexpression of the ERBB2 receptor tyrosine kinase and the mitochondrial inner membrane protein UCP2 occurs frequently in aggressive cancers with dysfunctional mitochondria. Overexpressed ERBB2 signals constitutively and elevated UCP2 can uncouple mitochondria and alleviate oxidative stress. However, the physiological contributions of UCP2 and ERBB2 at the low expression levels that are typical of most tissues, as well as the path to oncogenic deregulation, are poorly understood. We now show that ERBB2 directly controls UCP2 levels, both at low physiological levels and oncogenic overexpression. At low levels of receptor and UCP2, ligand stimulation creates a distinct temporal response pattern driven by the opposing forces of translational suppression of the exceptionally short lived UCP2 protein and a time delayed transcriptional up-regulation. The latter becomes dominant through constitutive signaling by overexpressed ERBB2, resulting in high levels of UCP2 that contribute mitochondrial uncoupling. By contrast, ligand stimulation of non-overexpressed ERBB2 transiently removes UCP2 and paradoxically reduces the mitochondrial membrane potential, oxygen consumption, and OXPHOS on a signaling time scale. However, neither the transporter activity nor down-regulation of already low UCP2 levels drive this reduction in mitochondrial activity. Instead, UCP2 is required to establish mitochondria that are capable of responding to ligand. UCP2 knockdown impairs proliferation at high glucose but its absence specifically impairs ligand-induced growth when glucose levels fluctuate. These findings demonstrate the ability of growth factor signaling to control oxidative phosphorylation on a signaling time scale and point toward a non-transporter role for low levels of UCP2 in establishing dynamic response capability.
Keywords: Glucose Metabolism; Mitochondria; Receptor Tyrosine Kinase; Signal Transduction; Uncoupling Proteins.
Figures




Similar articles
-
Increased activity of mitochondrial uncoupling protein 2 improves stress resistance in cultured endothelial cells exposed in vitro to high glucose levels.Am J Physiol Heart Circ Physiol. 2015 Jul 1;309(1):H147-56. doi: 10.1152/ajpheart.00759.2014. Epub 2015 Apr 24. Am J Physiol Heart Circ Physiol. 2015. PMID: 25910810
-
Mitochondrial retrograde signaling mediated by UCP2 inhibits cancer cell proliferation and tumorigenesis.Cancer Res. 2014 Jul 15;74(14):3971-82. doi: 10.1158/0008-5472.CAN-13-3383. Epub 2014 May 22. Cancer Res. 2014. PMID: 24853548
-
Transcriptional upregulation of mitochondrial uncoupling protein 2 protects against oxidative stress-associated neurogenic hypertension.Circ Res. 2009 Oct 23;105(9):886-96. doi: 10.1161/CIRCRESAHA.109.199018. Epub 2009 Sep 17. Circ Res. 2009. PMID: 19762685
-
New aspects of mitochondrial Uncoupling Proteins (UCPs) and their roles in tumorigenesis.Int J Mol Sci. 2011;12(8):5285-93. doi: 10.3390/ijms12085285. Epub 2011 Aug 17. Int J Mol Sci. 2011. PMID: 21954358 Free PMC article. Review.
-
Mitochondrial UCP2 in the central regulation of metabolism.Best Pract Res Clin Endocrinol Metab. 2014 Oct;28(5):757-64. doi: 10.1016/j.beem.2014.02.006. Epub 2014 Mar 7. Best Pract Res Clin Endocrinol Metab. 2014. PMID: 25256770 Review.
Cited by
-
Overexpression of ErbB2 renders breast cancer cells susceptible to 3-BrPA through the increased dissociation of hexokinase II from mitochondrial outer membrane.Oncol Lett. 2016 Feb;11(2):1567-1573. doi: 10.3892/ol.2015.4043. Epub 2015 Dec 21. Oncol Lett. 2016. PMID: 26893781 Free PMC article.
-
Therapeutic Target Analysis and Molecular Mechanism of Melatonin - Treated Leptin Resistance Induced Obesity: A Systematic Study of Network Pharmacology.Front Endocrinol (Lausanne). 2022 Jul 22;13:927576. doi: 10.3389/fendo.2022.927576. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35937803 Free PMC article.
-
Cerium Oxide Nanoparticle Administration to Skeletal Muscle Cells under Different Gravity and Radiation Conditions.ACS Appl Mater Interfaces. 2021 Sep 1;13(34):40200-40213. doi: 10.1021/acsami.1c14176. Epub 2021 Aug 19. ACS Appl Mater Interfaces. 2021. PMID: 34410709 Free PMC article.
-
Emerging role and the signaling pathways of uncoupling protein 2 in kidney diseases.Ren Fail. 2024 Dec;46(2):2381604. doi: 10.1080/0886022X.2024.2381604. Epub 2024 Aug 1. Ren Fail. 2024. PMID: 39090967 Free PMC article. Review.
-
UCP-2 inhibitor enhanced the efficacy of trastuzumab against HER2 positive breast cancer cells.Cancer Chemother Pharmacol. 2021 Oct;88(4):633-642. doi: 10.1007/s00280-021-04303-4. Epub 2021 Jun 19. Cancer Chemother Pharmacol. 2021. PMID: 34146128 Free PMC article.
References
-
- Winkler E., Klingenberg M. (1994) Effect of fatty acids on H+ transport activity of the reconstituted uncoupling protein. J. Biol. Chem. 269, 2508–2515 - PubMed
-
- Nedergaard J., Golozoubova V., Matthias A., Asadi A., Jacobsson A., Cannon B. (2001) UCP1. The only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochim. Biophys. Acta 1504, 82–106 - PubMed
-
- Divakaruni A. S., Brand M. D. (2011) The regulation and physiology of mitochondrial proton leak. Physiology 26, 192–205 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

