Dynamin 3: a new candidate tumor suppressor gene in hepatocellular carcinoma detected by triple combination array analysis
- PMID: 24143113
- PMCID: PMC3797647
- DOI: 10.2147/OTT.S51913
Dynamin 3: a new candidate tumor suppressor gene in hepatocellular carcinoma detected by triple combination array analysis
Abstract
Background: To identify genes associated with hepatocellular carcinoma (HCC) pathogenesis, we developed a triple combination array strategy comprising methylation, gene expression, and single nucleotide polymorphism (SNP) array analysis.
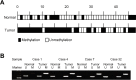
Methods: Surgical specimens obtained from a 68-year-old female HCC patient were analyzed by triple combination array, and identified Dynamin 3 (DNM3) as a candidate tumor suppressor gene in HCC. Subsequently, samples from 48 HCC patients were evaluated for DNM3 methylation and expression status using methylation specific polymerase chain reaction (PCR; MSP) and semi-quantitative reverse transcriptase (RT)-PCR, respectively. The relationship between clinicopathological factors and DNM3 methylation status was also investigated.
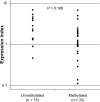
Results: DNM3 was shown to be hypermethylated (methylation value 0.879, range 0-1.0) in cancer tissue compared with adjacent normal tissue (0.213) by methylation array in the 68-year-old female patient. Expression arrays revealed decreased expression of DNM3 in cancerous tissue. SNP arrays revealed that the copy number of chromosome 1q24.3, in which DNM3 resides, was normal. MSP revealed hypermethylation of the DNM3 promoter region in 33 of 48 tumor samples. A trend toward decreased DNM3 expression was observed in patients with DNM3 promoter methylation (P = 0.189). Furthermore, patients with reduced expression of DNM3 in tumor tissues exhibited worse prognosis with decreased disease specific survival compared to patients without decreased expression (P = 0.014).
Conclusion: The present study indicates that a triple combination array strategy is an effective method to detect novel genes related to HCC. We propose that DNM3 is a tumor suppressor gene in HCC.
Keywords: DNM3; hepatocellular carcinoma; methylation; triple combination array.
Figures




Similar articles
-
Detection of doublecortin domain-containing 2 (DCDC2), a new candidate tumor suppressor gene of hepatocellular carcinoma, by triple combination array analysis.J Exp Clin Cancer Res. 2013 Sep 14;32(1):65. doi: 10.1186/1756-9966-32-65. J Exp Clin Cancer Res. 2013. PMID: 24034596 Free PMC article.
-
CCNJ detected by triple combination array analysis as a tumor-related gene of hepatocellular carcinoma.Int J Oncol. 2015 May;46(5):1963-70. doi: 10.3892/ijo.2015.2892. Epub 2015 Feb 11. Int J Oncol. 2015. PMID: 25672416
-
Protein tyrosine kinase 7: a hepatocellular carcinoma-related gene detected by triple-combination array.J Surg Res. 2015 May 15;195(2):444-53. doi: 10.1016/j.jss.2014.12.045. Epub 2014 Dec 31. J Surg Res. 2015. PMID: 25796105
-
Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis.Int J Oncol. 2013 Jul;43(1):88-94. doi: 10.3892/ijo.2013.1951. Epub 2013 May 20. Int J Oncol. 2013. PMID: 23695389
-
Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma.World J Gastroenterol. 2015 May 28;21(20):6317-28. doi: 10.3748/wjg.v21.i20.6317. World J Gastroenterol. 2015. PMID: 26034368 Free PMC article. Review.
Cited by
-
TBX15 rs98422, DNM3 rs1011731, RAD51B rs8017304, and rs2588809 Gene Polymorphisms and Associations With Pituitary Adenoma.In Vivo. 2021 Mar-Apr;35(2):815-826. doi: 10.21873/invivo.12322. In Vivo. 2021. PMID: 33622874 Free PMC article.
-
Identification of the collagen type 1 α 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma.BMC Cancer. 2014 Feb 19;14:108. doi: 10.1186/1471-2407-14-108. BMC Cancer. 2014. PMID: 24552139 Free PMC article.
-
Exosomal microRNA-23a-3p contributes to the progression of cholangiocarcinoma by interaction with Dynamin3.Bioengineered. 2022 Mar;13(3):6208-6221. doi: 10.1080/21655979.2022.2037249. Bioengineered. 2022. PMID: 35200104 Free PMC article.
-
siPRDX2-elevated DNM3 inhibits the proliferation and metastasis of colon cancer cells via AKT signaling pathway.Cancer Manag Res. 2019 Jun 28;11:5799-5811. doi: 10.2147/CMAR.S193805. eCollection 2019. Cancer Manag Res. 2019. PMID: 31388312 Free PMC article.
-
Prediction of Key Candidate Genes for Platinum Resistance in Ovarian Cancer.Int J Gen Med. 2021 Nov 16;14:8237-8248. doi: 10.2147/IJGM.S338044. eCollection 2021. Int J Gen Med. 2021. PMID: 34815697 Free PMC article.
References
-
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. - PubMed
-
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. - PubMed
-
- Livraghi T, Goldberg SN, Lazzaroni S, Meloni F, Solbiati L, Gazelle GS. Small hepatocellular carcinoma: treatment with radio-frequency ablation versus ethanol injection. Radiology. 1999;210(3):655–661. - PubMed
-
- Takayasu K, Arii S, Ikai I, et al. Liver Cancer Study Group of Japan Prospective cohort study of transarterial chemoembolization for unresectable hepatocellular carcinoma in 8510 patients. Gastroenterology. 2006;131(2):461–469. - PubMed
-
- Llovet JM, Ricci S, Mazzaferro V, et al. SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–390. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

