Indian signatures in the westernmost edge of the European Romani diaspora: new insight from mitogenomes
- PMID: 24143169
- PMCID: PMC3797067
- DOI: 10.1371/journal.pone.0075397
Indian signatures in the westernmost edge of the European Romani diaspora: new insight from mitogenomes
Abstract
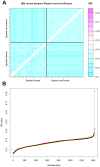
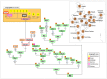
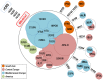
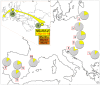
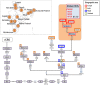
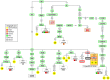
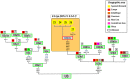
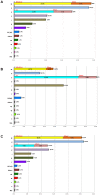
In agreement with historical documentation, several genetic studies have revealed ancestral links between the European Romani and India. The entire mitochondrial DNA (mtDNA) of 27 Spanish Romani was sequenced in order to shed further light on the origins of this population. The data were analyzed together with a large published dataset (mainly hypervariable region I [HVS-I] haplotypes) of Romani (N=1,353) and non-Romani worldwide populations (N>150,000). Analysis of mitogenomes allowed the characterization of various Romani-specific clades. M5a1b1a1 is the most distinctive European Romani haplogroup; it is present in all Romani groups at variable frequencies (with only sporadic findings in non-Romani) and represents 18% of their mtDNA pool. Its phylogeographic features indicate that M5a1b1a1 originated 1.5 thousand years ago (kya; 95% CI: 1.3-1.8) in a proto-Romani population living in Northwest India. U3 represents the most characteristic Romani haplogroup of European/Near Eastern origin (12.4%); it appears at dissimilar frequencies across the continent (Iberia: ≈ 31%; Eastern/Central Europe: ≈ 13%). All U3 mitogenomes of our Iberian Romani sample fall within a new sub-clade, U3b1c, which can be dated to 0.5 kya (95% CI: 0.3-0.7); therefore, signaling a lower bound for the founder event that followed admixture in Europe/Near East. Other minor European/Near Eastern haplogroups (e.g. H24, H88a) were also assimilated into the Romani by introgression with neighboring populations during their diaspora into Europe; yet some show a differentiation from the phylogenetically closest non-Romani counterpart. The phylogeny of Romani mitogenomes shows clear signatures of low effective population sizes and founder effects. Overall, these results are in good agreement with historical documentation, suggesting that cultural identity and relative isolation have allowed the Romani to preserve a distinctive mtDNA heritage, with some features linking them unequivocally to their ancestral Indian homeland.
Conflict of interest statement
Figures
References
-
- Liégois JP (2007) Roms en Europe: Éditions du Conseil de l'Europe.
-
- Ioviţă R, Schurr TG (2004) Reconstructing the origins and migrations of diasporic populations: the case of the European gypsies. Am Anthropol 106: 267–281.
-
- Fraser A (1995) The gypsies; Wiley-Blackwell, editor. Oxford UK: Blackwell Publishers.
-
- Turner RL (1984) The position of Romani in Indo-Aryan: Monographs.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

