The mitochondrial RNA landscape of Saccharomyces cerevisiae
- PMID: 24143261
- PMCID: PMC3797045
- DOI: 10.1371/journal.pone.0078105
The mitochondrial RNA landscape of Saccharomyces cerevisiae
Abstract
Mitochondria are essential organelles that harbor a reduced genome, and expression of that genome requires regulated metabolism of its transcriptome by nuclear-encoded proteins. Despite extensive investigation, a comprehensive map of the yeast mitochondrial transcriptome has not been developed and all of the RNA-metabolizing proteins have not been identified, both of which are prerequisites to elucidating the basic RNA biology of mitochondria. Here, we present a mitochondrial transcriptome map of the yeast S288C reference strain. Using RNAseq and bioinformatics, we show the expression level of all transcripts, revise all promoter, origin of replication, and tRNA annotations, and demonstrate for the first time the existence of alternative splicing, mirror RNAs, and a novel RNA processing site in yeast mitochondria. The transcriptome map has revealed new aspects of mitochondrial RNA biology and we expect it will serve as a valuable resource. As a complement to the map, we present our compilation of all known yeast nuclear-encoded ribonucleases (RNases), and a screen of this dataset for those that are imported into mitochondria. We sought to identify RNases that are refractory to recovery in traditional mitochondrial screens due to an essential function or eclipsed accumulation in another cellular compartment. Using this in silico approach, the essential RNase of the nuclear and cytoplasmic exosome, Dis3p, emerges as a strong candidate. Bioinformatics and in vivo analyses show that Dis3p has a conserved and functional mitochondrial-targeting signal (MTS). A clean and marker-less chromosomal deletion of the Dis3p MTS results in a defect in the decay of intron and mirror RNAs, thus revealing a role for Dis3p in mitochondrial RNA decay.
Conflict of interest statement
Figures





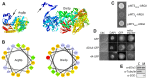
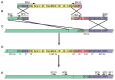
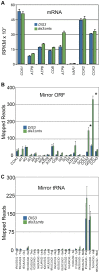

References
-
- Ephrussi B (1949) Action de l’acriflavine sur les levures. In: Unités Biologiques Douées de Continuité Génétiqué. du Paris: Centre Nat Rech Sci: 165-180.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

