An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice
- PMID: 24143807
- PMCID: PMC3850199
- DOI: 10.1104/pp.113.224881
An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice
Abstract
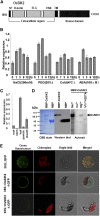
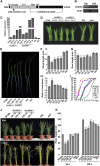
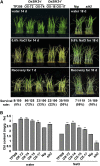
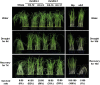
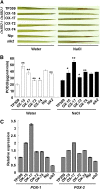
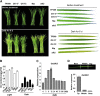
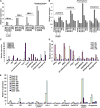
Receptor-like kinases play important roles in plant development and defense responses; however, their functions in other processes remain unclear. Here, we report that OsSIK2, an S-domain receptor-like kinase from rice (Oryza sativa), is involved in abiotic stress and the senescence process. OsSIK2 is a plasma membrane-localized protein with kinase activity in the presence of Mn(2+). OsSIK2 is expressed mainly in rice leaf and sheath and can be induced by NaCl, drought, cold, dark, and abscisic acid treatment. Transgenic plants overexpressing OsSIK2 and mutant sik2 exhibit enhanced and reduced tolerance to salt and drought stress, respectively, compared with the controls. Interestingly, a truncated version of OsSIK2 without most of the extracellular region confers higher salt tolerance than the full-length OsSIK2, likely through the activation of different sets of downstream genes. Moreover, seedlings of OsSIK2-overexpressing transgenic plants exhibit early leaf development and a delayed dark-induced senescence phenotype, while mutant sik2 shows the opposite phenotype. The downstream PR-related genes specifically up-regulated by full-length OsSIK2 or the DREB-like genes solely enhanced by truncated OsSIK2 are all induced by salt, drought, and dark treatments. These results indicate that OsSIK2 may integrate stress signals into a developmental program for better adaptive growth under unfavorable conditions. Manipulation of OsSIK2 should facilitate the improvement of production in rice and other crops.
Figures

Similar articles
-
OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes.Plant Physiol. 2013 Mar;161(3):1202-16. doi: 10.1104/pp.112.205385. Epub 2013 Jan 7. Plant Physiol. 2013. PMID: 23296688 Free PMC article.
-
The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice.BMC Plant Biol. 2008 Apr 28;8:49. doi: 10.1186/1471-2229-8-49. BMC Plant Biol. 2008. PMID: 18442365 Free PMC article.
-
OsSIDP366, a DUF1644 gene, positively regulates responses to drought and salt stresses in rice.J Integr Plant Biol. 2016 May;58(5):492-502. doi: 10.1111/jipb.12376. Epub 2015 Sep 25. J Integr Plant Biol. 2016. PMID: 26172270
-
Physiological and Molecular Mechanisms of Rice Tolerance to Salt and Drought Stress: Advances and Future Directions.Int J Mol Sci. 2024 Aug 29;25(17):9404. doi: 10.3390/ijms25179404. Int J Mol Sci. 2024. PMID: 39273349 Free PMC article. Review.
-
The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants.Plant Cell Rep. 2017 Feb;36(2):235-242. doi: 10.1007/s00299-016-2084-x. Epub 2016 Dec 8. Plant Cell Rep. 2017. PMID: 27933379 Review.
Cited by
-
Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants.Front Plant Sci. 2015 Feb 18;6:84. doi: 10.3389/fpls.2015.00084. eCollection 2015. Front Plant Sci. 2015. PMID: 25741357 Free PMC article. Review.
-
Identification and Functional Analysis of Key Autophosphorylation Residues of Arabidopsis Senescence Associated Receptor-like Kinase.Int J Mol Sci. 2022 Aug 9;23(16):8873. doi: 10.3390/ijms23168873. Int J Mol Sci. 2022. PMID: 36012141 Free PMC article.
-
Ectopic Expression of GsSRK in Medicago sativa Reveals Its Involvement in Plant Architecture and Salt Stress Responses.Front Plant Sci. 2018 Feb 22;9:226. doi: 10.3389/fpls.2018.00226. eCollection 2018. Front Plant Sci. 2018. PMID: 29520291 Free PMC article.
-
Plasma membrane receptor-like kinase leaf panicle 2 acts downstream of the DROUGHT AND SALT TOLERANCE transcription factor to regulate drought sensitivity in rice.J Exp Bot. 2015 Jan;66(1):271-81. doi: 10.1093/jxb/eru417. Epub 2014 Nov 10. J Exp Bot. 2015. PMID: 25385766 Free PMC article.
-
Understanding the response in Pugionium cornutum (L.) Gaertn. seedling leaves under drought stress using transcriptome and proteome integrated analysis.PeerJ. 2023 Apr 3;11:e15165. doi: 10.7717/peerj.15165. eCollection 2023. PeerJ. 2023. PMID: 37033724 Free PMC article.
References
-
- Balazadeh S, Riaño-Pachón DM, Mueller-Roeber B. (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttg) (Suppl 1) 10: 63–75 - PubMed
-
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 - PubMed
-
- Becraft PW, Stinard PS, McCarty DR. (1996) CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273: 1406–1409 - PubMed
-
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

