The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice
- PMID: 24144792
- PMCID: PMC3850201
- DOI: 10.1104/pp.113.224527
The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice
Abstract
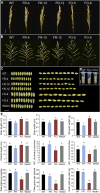
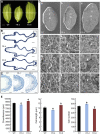
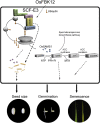
Leaf senescence is related to the grain-filling rate and grain weight in cereals. Many components involved in senescence regulation at either the genetic or physiological level are known. However, less is known about molecular regulation mechanisms. Here, we report that OsFBK12 (an F-box protein containing a Kelch repeat motif) interacts with S-ADENOSYL-l-METHIONINE SYNTHETASE1 (SAMS1) to regulate leaf senescence and seed size as well as grain number in rice (Oryza sativa). Yeast two-hybrid, pull-down, and bimolecular fluorescence complementation assays indicate that OsFBK12 interacts with Oryza sativa S-PHASE KINASE-ASSOCIATED PROTEIN1-LIKE PROTEIN and with OsSAMS1. Biochemical and physiological data showed that OsFBK12 targets OsSAMS1 for degradation. OsFBK12-RNA interference lines and OsSAMS1 overexpression lines showed increased ethylene levels, while OsFBK12-OX lines and OsSAMS1-RNA interference plants exhibited decreased ethylene. Phenotypically, overexpression of OsFBK12 led to a delay in leaf senescence and germination and increased seed size, whereas knockdown lines of either OsFBK12 or OsSAMS1 promoted the senescence program. Our results suggest that OsFBK12 is involved in the 26S proteasome pathway by interacting with Oryza sativa S-PHASE KINASE-ASSOCIATED PROTEIN1-LIKE PROTEIN and that it targets the substrate OsSAMS1 for degradation, triggering changes in ethylene levels for the regulation of leaf senescence and grain size. These data have potential applications in the molecular breeding of rice.
Figures




Similar articles
-
OsLCD3 interacts with OsSAMS1 to regulate grain size via ethylene/polyamine homeostasis control.Plant J. 2024 Jul;119(2):705-719. doi: 10.1111/tpj.16788. Epub 2024 May 4. Plant J. 2024. PMID: 38703081
-
Knockdown of SAMS genes encoding S-adenosyl-l-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice.J Plant Physiol. 2011 Oct 15;168(15):1837-43. doi: 10.1016/j.jplph.2011.05.020. Epub 2011 Jul 14. J Plant Physiol. 2011. PMID: 21757254
-
A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis.Plant Physiol. 2017 Jul;174(3):1747-1763. doi: 10.1104/pp.17.00542. Epub 2017 May 12. Plant Physiol. 2017. PMID: 28500268 Free PMC article.
-
Current Understanding of Leaf Senescence in Rice.Int J Mol Sci. 2021 Apr 26;22(9):4515. doi: 10.3390/ijms22094515. Int J Mol Sci. 2021. PMID: 33925978 Free PMC article. Review.
-
Genetic Dissection of Leaf Senescence in Rice.Int J Mol Sci. 2017 Dec 11;18(12):2686. doi: 10.3390/ijms18122686. Int J Mol Sci. 2017. PMID: 29232920 Free PMC article. Review.
Cited by
-
S-Adenosylmethionine Synthetase 3 Is Important for Pollen Tube Growth.Plant Physiol. 2016 Sep;172(1):244-53. doi: 10.1104/pp.16.00774. Epub 2016 Aug 1. Plant Physiol. 2016. PMID: 27482079 Free PMC article.
-
Du13 encodes a C2 H2 zinc-finger protein that regulates Wxb pre-mRNA splicing and microRNA biogenesis in rice endosperm.Plant Biotechnol J. 2022 Jul;20(7):1387-1401. doi: 10.1111/pbi.13821. Epub 2022 May 13. Plant Biotechnol J. 2022. PMID: 35560858 Free PMC article.
-
Genetic Localization and Homologous Genes Mining for Barley Grain Size.Int J Mol Sci. 2023 Mar 3;24(5):4932. doi: 10.3390/ijms24054932. Int J Mol Sci. 2023. PMID: 36902360 Free PMC article. Review.
-
Impaired Function of the Calcium-Dependent Protein Kinase, OsCPK12, Leads to Early Senescence in Rice (Oryza sativa L.).Front Plant Sci. 2019 Feb 4;10:52. doi: 10.3389/fpls.2019.00052. eCollection 2019. Front Plant Sci. 2019. PMID: 30778363 Free PMC article.
-
Wheat F-Box Protein Gene TaFBA1 Is Involved in Plant Tolerance to Heat Stress.Front Plant Sci. 2018 Apr 24;9:521. doi: 10.3389/fpls.2018.00521. eCollection 2018. Front Plant Sci. 2018. PMID: 29740462 Free PMC article.
References
-
- Chen N, Xu Y, Wang X, Du C, Du J, Yuan M, Xu Z, Chong K. (2011) OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ 34: 52–64 - PubMed
-
- Comcepcion M, Lizada C, Yang SF. (1979) A simple and sensitive assay for 1-aminocyclopropane-1-carboxylic acid. Anal Biochem 100: 140–145 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

