Landscape of somatic mutations and clonal evolution in mantle cell lymphoma
- PMID: 24145436
- PMCID: PMC3831489
- DOI: 10.1073/pnas.1314608110
Landscape of somatic mutations and clonal evolution in mantle cell lymphoma
Abstract
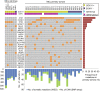
Mantle cell lymphoma (MCL) is an aggressive tumor, but a subset of patients may follow an indolent clinical course. To understand the mechanisms underlying this biological heterogeneity, we performed whole-genome and/or whole-exome sequencing on 29 MCL cases and their respective matched normal DNA, as well as 6 MCL cell lines. Recurrently mutated genes were investigated by targeted sequencing in an independent cohort of 172 MCL patients. We identified 25 significantly mutated genes, including known drivers such as ataxia-telangectasia mutated (ATM), cyclin D1 (CCND1), and the tumor suppressor TP53; mutated genes encoding the anti-apoptotic protein BIRC3 and Toll-like receptor 2 (TLR2); and the chromatin modifiers WHSC1, MLL2, and MEF2B. We also found NOTCH2 mutations as an alternative phenomenon to NOTCH1 mutations in aggressive tumors with a dismal prognosis. Analysis of two simultaneous or subsequent MCL samples by whole-genome/whole-exome (n = 8) or targeted (n = 19) sequencing revealed subclonal heterogeneity at diagnosis in samples from different topographic sites and modulation of the initial mutational profile at the progression of the disease. Some mutations were predominantly clonal or subclonal, indicating an early or late event in tumor evolution, respectively. Our study identifies molecular mechanisms contributing to MCL pathogenesis and offers potential targets for therapeutic intervention.
Keywords: cancer genetics; cancer heterogeneity; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Rosenwald A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. - PubMed
-
- Vose JM. Mantle cell lymphoma: 2012 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2012;87(6):604–609. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

