Biofilm matrix and its regulation in Pseudomonas aeruginosa
- PMID: 24145749
- PMCID: PMC3821654
- DOI: 10.3390/ijms141020983
Biofilm matrix and its regulation in Pseudomonas aeruginosa
Abstract
Biofilms are communities of microorganisms embedded in extracellular polymeric substances (EPS) matrix. Bacteria in biofilms demonstrate distinct features from their free-living planktonic counterparts, such as different physiology and high resistance to immune system and antibiotics that render biofilm a source of chronic and persistent infections. A deeper understanding of biofilms will ultimately provide insights into the development of alternative treatment for biofilm infections. The opportunistic pathogen Pseudomonas aeruginosa, a model bacterium for biofilm research, is notorious for its ability to cause chronic infections by its high level of drug resistance involving the formation of biofilms. In this review, we summarize recent advances in biofilm formation, focusing on the biofilm matrix and its regulation in P. aeruginosa, aiming to provide resources for the understanding and control of bacterial biofilms.
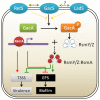
Figures



References
-
- Davies D.G., Parsek M.R., Pearson J.P., Iglewski B.H., Costerton J.W., Greenberg E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. - PubMed
-
- Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. 2004;2:95–108. - PubMed
-
- Sutherland I.W. The biofilm matrix—An immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. - PubMed
-
- Branda S.S., Vik S., Friedman L., Kolter R. Biofilms: The matrix revisited. Trends Microbiol. 2005;13:20–26. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

