A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli
- PMID: 24146630
- PMCID: PMC3798268
- DOI: 10.1371/journal.pgen.1003865
A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli
Erratum in
- PLoS Genet. 2013 Oct;9(10). doi:10.1371/annotation/1e9bcb70-265a-4383-abf4-3466d144d56e. Liu, Zhangzhi [corrected to Liu, Zhanzhi]
Abstract
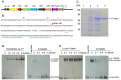
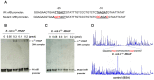
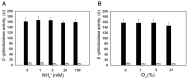
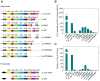
Most biological nitrogen fixation is catalyzed by molybdenum-dependent nitrogenase, an enzyme complex comprising two component proteins that contains three different metalloclusters. Diazotrophs contain a common core of nitrogen fixation nif genes that encode the structural subunits of the enzyme and components required to synthesize the metalloclusters. However, the complement of nif genes required to enable diazotrophic growth varies significantly amongst nitrogen fixing bacteria and archaea. In this study, we identified a minimal nif gene cluster consisting of nine nif genes in the genome of Paenibacillus sp. WLY78, a gram-positive, facultative anaerobe isolated from the rhizosphere of bamboo. We demonstrate that the nif genes in this organism are organized as an operon comprising nifB, nifH, nifD, nifK, nifE, nifN, nifX, hesA and nifV and that the nif cluster is under the control of a σ(70) (σ(A))-dependent promoter located upstream of nifB. To investigate genetic requirements for diazotrophy, we transferred the Paenibacillus nif cluster to Escherichia coli. The minimal nif gene cluster enables synthesis of catalytically active nitrogenase in this host, when expressed either from the native nifB promoter or from the T7 promoter. Deletion analysis indicates that in addition to the core nif genes, hesA plays an important role in nitrogen fixation and is responsive to the availability of molybdenum. Whereas nif transcription in Paenibacillus is regulated in response to nitrogen availability and by the external oxygen concentration, transcription from the nifB promoter is constitutive in E. coli, indicating that negative regulation of nif transcription is bypassed in the heterologous host. This study demonstrates the potential for engineering nitrogen fixation in a non-nitrogen fixing organism with a minimum set of nine nif genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Falkowski PG (1997) Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature 387: 272–275.
-
- Merrick M, Dixon R (1984) Why don't plants fix nitrogen? Trends Biotechnol 2: 162–166.
-
- Beatty PH, Good AG (2011) Future prospects for cereals that fix nitrogen. Science 333: 416–417. - PubMed
-
- Rubio LM, Ludden PW (2008) Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol 62: 93–111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

