A novel branched chain amino acids responsive transcriptional regulator, BCARR, negatively acts on the proteolytic system in Lactobacillus helveticus
- PMID: 24146802
- PMCID: PMC3795697
- DOI: 10.1371/journal.pone.0075976
A novel branched chain amino acids responsive transcriptional regulator, BCARR, negatively acts on the proteolytic system in Lactobacillus helveticus
Abstract
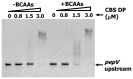
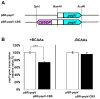
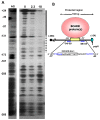
Transcriptional negative regulation of the proteolytic system of Lactobacillus helveticus CM4 in response to amino acids seems to be very important for the control of antihypertensive peptide production; however, it remains poorly understood. A 26-kDa protein with N-terminal cystathionine β-synthase domains (CBS domain protein), which seems to be involved in the regulatory system, was purified by using a DNA-sepharose bound 300-bp DNA fragment corresponding to the upstream regions of the six proteolytic genes that are down-regulated by amino acids. The CBS domain protein bound to a DNA fragment corresponding to the region upstream of the pepV gene in response to branched chain amino acids (BCAAs). The expression of the pepV gene in Escherichia coli grown in BCAA-enriched medium was repressed when the CBS domain protein was co-expressed. These results reveal that the CBS domain protein acts as a novel type of BCAA-responsive transcriptional regulator (BCARR) in L. helveticus. From comparative analysis of the promoter regions of the six proteolysis genes, a palindromic AT-rich motif, 5'-AAAAANNCTWTTATT-3', was predicted as the consensus DNA motif for the BCARR protein binding. Footprint analysis using the pepV promotor region and gel shift analyses with the corresponding short DNA fragments strongly suggested that the BCARR protein binds adjacent to the pepV promoter region and affects the transcription level of the pepV gene in the presence of BCAAs. Homology search analysis of the C-terminal region of the BCARR protein suggested the existence of a unique βαββαβ fold structure that has been reported in a variety of ACT (aspartate kinase-chorismate mutase-tyrA) domain proteins for sensing amino acids. These results also suggest that the sensing of BCAAs by the ACT domain might promote the binding of the BCARR to DNA sequences upstream of proteolysis genes, which affects the gene expression of the proteolytic system in L. helveticus.
Conflict of interest statement
Figures






Similar articles
-
Genome-wide motif predictions of BCARR-box in the amino-acid repressed genes of Lactobacillus helveticus CM4.BMC Microbiol. 2017 Dec 2;17(1):224. doi: 10.1186/s12866-017-1125-0. BMC Microbiol. 2017. PMID: 29197337 Free PMC article.
-
Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis.Mol Microbiol. 2001 Jun;40(5):1227-39. doi: 10.1046/j.1365-2958.2001.02470.x. Mol Microbiol. 2001. PMID: 11401725
-
Comparative analysis of proteolytic enzymes need for processing of antihypertensive peptides between Lactobacillus helveticus CM4 and DPC4571.J Biosci Bioeng. 2013 Mar;115(3):246-52. doi: 10.1016/j.jbiosc.2012.09.020. Epub 2012 Nov 20. J Biosci Bioeng. 2013. PMID: 23182500
-
FNR and its role in oxygen-regulated gene expression in Escherichia coli.FEMS Microbiol Rev. 1990 Aug;6(4):399-428. doi: 10.1111/j.1574-6968.1990.tb04109.x. FEMS Microbiol Rev. 1990. PMID: 2248796 Review.
-
Oxygen regulated gene expression in Escherichia coli: control of anaerobic respiration by the FNR protein.Antonie Van Leeuwenhoek. 1991 Feb;59(2):65-76. doi: 10.1007/BF00445650. Antonie Van Leeuwenhoek. 1991. PMID: 1854188 Review.
Cited by
-
YebC, a putative transcriptional factor involved in the regulation of the proteolytic system of Lactobacillus.Sci Rep. 2017 Aug 17;7(1):8579. doi: 10.1038/s41598-017-09124-1. Sci Rep. 2017. PMID: 28819300 Free PMC article.
-
Pathway and Production Differences in Branched-Chain Hydroxy Acids as Bioactive Metabolites in Limosilactobacillus fermentum, Ligilactobacillus salivarius, and Latilactobacillus sakei.Int J Mol Sci. 2024 Sep 20;25(18):10112. doi: 10.3390/ijms251810112. Int J Mol Sci. 2024. PMID: 39337595 Free PMC article.
-
Use of Mass Spectrometry to Profile Peptides in Whey Protein Isolate Medium Fermented by Lactobacillus helveticus LH-2 and Lactobacillus acidophilus La-5.Front Nutr. 2019 Oct 15;6:152. doi: 10.3389/fnut.2019.00152. eCollection 2019. Front Nutr. 2019. PMID: 31681785 Free PMC article.
-
In Silico Comparative Genomic Analysis Revealed a Highly Conserved Proteolytic System in Lactobacillus delbrueckii.Int J Mol Sci. 2023 Jul 11;24(14):11309. doi: 10.3390/ijms241411309. Int J Mol Sci. 2023. PMID: 37511069 Free PMC article.
-
Genome-wide motif predictions of BCARR-box in the amino-acid repressed genes of Lactobacillus helveticus CM4.BMC Microbiol. 2017 Dec 2;17(1):224. doi: 10.1186/s12866-017-1125-0. BMC Microbiol. 2017. PMID: 29197337 Free PMC article.
References
-
- Slattery L, O’Callaghan J, Fitzgerald G, Beresford T, Ross R (2010) Invited review: Lactobacillus helveticus–A thermophilic dairy starter related to gut bacteria. Journal of dairy science 93: 4435–4454. - PubMed
-
- Sneath PH, Mair NS, Sharpe ME, Holt JG (1986) Bergey’s manual of systematic bacteriology. Volume 2: Williams & Wilkins.
-
- Sadat-Mekmene L, Genay M, Atlan D, Lortal S, Gagnaire V (2011) Original features of cell-envelope proteinases of Lactobacillus helveticus. A review. International Journal of Food Microbiology 146: 1–13. - PubMed
-
- Yamamoto N, Akino A, Takano T (1994) Antihypertensive effects of different kinds of fermented milk in spontaneously hypertensive rats. Bioscience, biotechnology, and biochemistry 58: 776–778.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

