A modular lentiviral and retroviral construction system to rapidly generate vectors for gene expression and gene knockdown in vitro and in vivo
- PMID: 24146852
- PMCID: PMC3795761
- DOI: 10.1371/journal.pone.0076279
A modular lentiviral and retroviral construction system to rapidly generate vectors for gene expression and gene knockdown in vitro and in vivo
Abstract
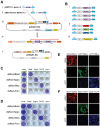
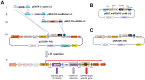
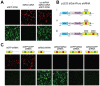
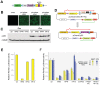
The ability to express exogenous cDNAs while suppressing endogenous genes via RNAi represents an extremely powerful research tool with the most efficient non-transient approach being accomplished through stable viral vector integration. Unfortunately, since traditional restriction enzyme based methods for constructing such vectors are sequence dependent, their construction is often difficult and not amenable to mass production. Here we describe a non-sequence dependent Gateway recombination cloning system for the rapid production of novel lentiviral (pLEG) and retroviral (pREG) vectors. Using this system to recombine 3 or 4 modular plasmid components it is possible to generate viral vectors expressing cDNAs with or without inhibitory RNAs (shRNAmirs). In addition, we demonstrate a method to rapidly produce and triage novel shRNAmirs for use with this system. Once strong candidate shRNAmirs have been identified they may be linked together in tandem to knockdown expression of multiple targets simultaneously or to improve the knockdown of a single target. Here we demonstrate that these recombinant vectors are able to express cDNA and effectively knockdown protein expression using both cell culture and animal model systems.
Conflict of interest statement
Figures
Similar articles
-
Construction of Modular Lentiviral Vectors for Effective Gene Expression and Knockdown.Methods Mol Biol. 2016;1448:3-21. doi: 10.1007/978-1-4939-3753-0_1. Methods Mol Biol. 2016. PMID: 27317169
-
DNA vector-based RNA interference to study gene function in cancer.J Vis Exp. 2012 Jun 4;(64):e4129. doi: 10.3791/4129. J Vis Exp. 2012. PMID: 22710444 Free PMC article.
-
Multipurpose modular lentiviral vectors for RNA interference and transgene expression.Mol Biol Rep. 2010 Jul;37(6):2863-70. doi: 10.1007/s11033-009-9840-8. Epub 2009 Oct 2. Mol Biol Rep. 2010. PMID: 19798586
-
Library selection approaches to engineering enhanced retroviral and lentiviral vectors.Comb Chem High Throughput Screen. 2008 Feb;11(2):111-7. doi: 10.2174/138620708783744444. Comb Chem High Throughput Screen. 2008. PMID: 18336204 Review.
-
Safety considerations in vector development.Somat Cell Mol Genet. 2001 Nov;26(1-6):147-58. doi: 10.1023/a:1021082815013. Somat Cell Mol Genet. 2001. PMID: 12465466 Review.
Cited by
-
Gram scale preparation of clozapine N-oxide (CNO), a synthetic small molecule actuator for muscarinic acetylcholine DREADDs.MethodsX. 2018 Mar 23;5:257-267. doi: 10.1016/j.mex.2018.03.003. eCollection 2018. MethodsX. 2018. PMID: 30038895 Free PMC article.
-
The inhibitor of kappa B kinase-epsilon regulates MMP-3 expression levels and can promote lung metastasis.Oncogenesis. 2014 Aug 18;3(8):e116. doi: 10.1038/oncsis.2014.28. Oncogenesis. 2014. PMID: 25133483 Free PMC article.
-
RAS transformation requires CUX1-dependent repair of oxidative DNA damage.PLoS Biol. 2014 Mar 11;12(3):e1001807. doi: 10.1371/journal.pbio.1001807. eCollection 2014 Mar. PLoS Biol. 2014. PMID: 24618719 Free PMC article.
-
Ras effector mutant expression suggest a negative regulator inhibits lung tumor formation.PLoS One. 2014 Jan 28;9(1):e84745. doi: 10.1371/journal.pone.0084745. eCollection 2014. PLoS One. 2014. PMID: 24489653 Free PMC article.
-
Caspase 9b Drives Cellular Transformation, Lung Inflammation, and Lung Tumorigenesis.Mol Cancer Res. 2022 Aug 5;20(8):1284-1294. doi: 10.1158/1541-7786.MCR-21-0905. Mol Cancer Res. 2022. PMID: 35412615 Free PMC article.
References
-
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, et al. (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272: 263–267. - PubMed
-
- Fewell GD, Schmitt K (2006) Vector-based RNAi approaches for stable, inducible and genome-wide screens. Drug Discov Today 11: 975–982. - PubMed
-
- Sakuma T, Barry MA, Ikeda Y (2012) Lentiviral vectors: basic to translational. Biochem J 443: 603–618. - PubMed
-
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

