Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes
- PMID: 24146880
- PMCID: PMC3797821
- DOI: 10.1371/journal.pone.0076511
Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes
Abstract
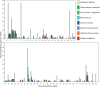
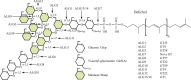
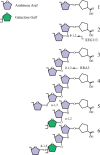
The Archaeplastida consists of three lineages, Rhodophyta, Virideplantae and Glaucophyta. The extracellular matrix of most members of the Rhodophyta and Viridiplantae consists of carbohydrate-based or a highly glycosylated protein-based cell wall while the Glaucophyte covering is poorly resolved. In order to elucidate possible evolutionary links between the three advanced lineages in Archaeplastida, a genomic analysis was initiated. Fully sequenced genomes from the Rhodophyta and Virideplantae and the well-defined CAZy database on glycosyltransferases were included in the analysis. The number of glycosyltransferases found in the Rhodophyta and Chlorophyta are generally much lower then in land plants (Embryophyta). Three specific features exhibited by land plants increase the number of glycosyltransferases in their genomes: (1) cell wall biosynthesis, the more complex land plant cell walls require a larger number of glycosyltransferases for biosynthesis, (2) a richer set of protein glycosylation, and (3) glycosylation of secondary metabolites, demonstrated by a large proportion of family GT1 being involved in secondary metabolite biosynthesis. In a comparative analysis of polysaccharide biosynthesis amongst the taxa of this study, clear distinctions or similarities were observed in (1) N-linked protein glycosylation, i.e., Chlorophyta has different mannosylation and glucosylation patterns, (2) GPI anchor biosynthesis, which is apparently missing in the Rhodophyta and truncated in the Chlorophyta, (3) cell wall biosynthesis, where the land plants have unique cell wall related polymers not found in green and red algae, and (4) O-linked glycosylation where comprehensive orthology was observed in glycosylation between the Chlorophyta and land plants but not between the target proteins.
Conflict of interest statement
Figures

Similar articles
-
Identification, classification, and evolution of putative xylosyltransferases from algae.Protoplasma. 2019 Jul;256(4):1119-1132. doi: 10.1007/s00709-019-01358-2. Epub 2019 Apr 3. Protoplasma. 2019. PMID: 30941581
-
Evidence for land plant cell wall biosynthetic mechanisms in charophyte green algae.Ann Bot. 2014 Oct;114(6):1217-36. doi: 10.1093/aob/mcu171. Epub 2014 Sep 9. Ann Bot. 2014. PMID: 25204387 Free PMC article.
-
Evolutionary and genomic analysis of the caleosin/peroxygenase (CLO/PXG) gene/protein families in the Viridiplantae.PLoS One. 2018 May 17;13(5):e0196669. doi: 10.1371/journal.pone.0196669. eCollection 2018. PLoS One. 2018. PMID: 29771926 Free PMC article.
-
What Happened to the Phycobilisome?Biomolecules. 2019 Nov 19;9(11):748. doi: 10.3390/biom9110748. Biomolecules. 2019. PMID: 31752285 Free PMC article. Review.
-
The Land-Sea Connection: Insights Into the Plant Lineage from a Green Algal Perspective.Annu Rev Plant Biol. 2022 May 20;73:585-616. doi: 10.1146/annurev-arplant-071921-100530. Epub 2022 Mar 8. Annu Rev Plant Biol. 2022. PMID: 35259927 Review.
Cited by
-
Genomes of early-diverging streptophyte algae shed light on plant terrestrialization.Nat Plants. 2020 Feb;6(2):95-106. doi: 10.1038/s41477-019-0560-3. Epub 2019 Dec 16. Nat Plants. 2020. PMID: 31844283 Free PMC article.
-
Identification, classification, and evolution of putative xylosyltransferases from algae.Protoplasma. 2019 Jul;256(4):1119-1132. doi: 10.1007/s00709-019-01358-2. Epub 2019 Apr 3. Protoplasma. 2019. PMID: 30941581
-
Life cycle and functional genomics of the unicellular red alga Galdieria for elucidating algal and plant evolution and industrial use.Proc Natl Acad Sci U S A. 2022 Oct 11;119(41):e2210665119. doi: 10.1073/pnas.2210665119. Epub 2022 Oct 4. Proc Natl Acad Sci U S A. 2022. PMID: 36194630 Free PMC article.
-
The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.Nat Commun. 2022 Jun 8;13(1):3314. doi: 10.1038/s41467-022-31048-2. Nat Commun. 2022. PMID: 35676258 Free PMC article.
-
Insights into the Evolution of Hydroxyproline-Rich Glycoproteins from 1000 Plant Transcriptomes.Plant Physiol. 2017 Jun;174(2):904-921. doi: 10.1104/pp.17.00295. Epub 2017 Apr 26. Plant Physiol. 2017. PMID: 28446636 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

