High-throughput sequencing of islet-infiltrating memory CD4+ T cells reveals a similar pattern of TCR Vβ usage in prediabetic and diabetic NOD mice
- PMID: 24146886
- PMCID: PMC3798422
- DOI: 10.1371/journal.pone.0076546
High-throughput sequencing of islet-infiltrating memory CD4+ T cells reveals a similar pattern of TCR Vβ usage in prediabetic and diabetic NOD mice
Abstract
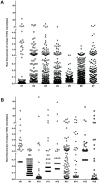
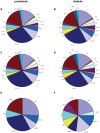
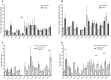
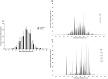
Autoreactive memory CD4(+) T cells play a critical role in the development of type 1 diabetes, but it is not yet known how the clonotypic composition and TCRβ repertoire of the memory CD4(+) T cell compartment changes during the transition from prediabetes to diabetes. In this study, we used high-throughput sequencing to analyze the TCRβ repertoire of sorted islet-infiltrating memory CD4(+)CD44(high) T cells in 10-week-old prediabetic and recently diabetic NOD mice. We show that most clonotypes of islet-infiltrating CD4(+)CD44(high) T cells were rare, but high-frequency clonotypes were significantly more common in diabetic than in prediabetic mice. Moreover, although the CD4(+)CD44(high) TCRβ repertoires were highly diverse at both stages of disease development, dominant use of TRBV1 (Vβ2), TRBV13-3 (Vβ8.1), and TRBV19 (Vβ6) was evident in both prediabetic and diabetic mice. Our findings strongly suggest that therapeutic targeting of cells specifically expressing the dominant TCRβ might reduce pancreatic infiltration in prediabetic mice and attenuate the progression to diabetes.
Conflict of interest statement
Figures



Similar articles
-
High-throughput sequencing reveals restricted TCR Vβ usage and public TCRβ clonotypes among pancreatic lymph node memory CD4(+) T cells and their involvement in autoimmune diabetes.Mol Immunol. 2016 Jun;74:82-95. doi: 10.1016/j.molimm.2016.04.013. Epub 2016 May 6. Mol Immunol. 2016. PMID: 27161799 Free PMC article.
-
Islet-associated T-cell receptor-β CDR sequence repertoire in prediabetic NOD mice reveals antigen-driven T-cell expansion and shared usage of VβJβ TCR chains.Mol Immunol. 2015 Mar;64(1):127-35. doi: 10.1016/j.molimm.2014.11.009. Epub 2014 Dec 3. Mol Immunol. 2015. PMID: 25480393
-
Autoreactive effector/memory CD4+ and CD8+ T cells infiltrating grafted and endogenous islets in diabetic NOD mice exhibit similar T cell receptor usage.PLoS One. 2012;7(12):e52054. doi: 10.1371/journal.pone.0052054. Epub 2012 Dec 14. PLoS One. 2012. PMID: 23251685 Free PMC article.
-
Type I diabetes mellitus: a predictable autoimmune disease with interindividual variation in the rate of beta cell destruction.Clin Immunol Immunopathol. 1989 Jan;50(1 Pt 2):S85-95. doi: 10.1016/0090-1229(89)90115-3. Clin Immunol Immunopathol. 1989. PMID: 2642771 Review.
-
Control of type 1 autoimmune diabetes by naturally occurring CD4+CD25+ regulatory T lymphocytes in neonatal NOD mice.Ann N Y Acad Sci. 2005 Jun;1051:72-87. doi: 10.1196/annals.1361.048. Ann N Y Acad Sci. 2005. PMID: 16126946 Review.
Cited by
-
In Vivo Enrichment of Diabetogenic T Cells.Diabetes. 2017 Aug;66(8):2220-2229. doi: 10.2337/db16-0946. Epub 2017 Apr 10. Diabetes. 2017. PMID: 28396510 Free PMC article.
-
RepSeq Data Representativeness and Robustness Assessment by Shannon Entropy.Front Immunol. 2018 May 15;9:1038. doi: 10.3389/fimmu.2018.01038. eCollection 2018. Front Immunol. 2018. PMID: 29868003 Free PMC article.
-
High-throughput T cell receptor sequencing identifies clonally expanded CD8+ T cell populations in alopecia areata.JCI Insight. 2018 Oct 4;3(19):e121949. doi: 10.1172/jci.insight.121949. JCI Insight. 2018. PMID: 30282836 Free PMC article.
-
T-cell receptor repertoire variation may be associated with type 2 diabetes mellitus in humans.Diabetes Metab Res Rev. 2016 Mar;32(3):297-307. doi: 10.1002/dmrr.2720. Epub 2016 Jan 13. Diabetes Metab Res Rev. 2016. PMID: 26408818 Free PMC article.
-
CD8(+) T-cell pathogenicity in Rasmussen encephalitis elucidated by large-scale T-cell receptor sequencing.Nat Commun. 2016 Apr 4;7:11153. doi: 10.1038/ncomms11153. Nat Commun. 2016. PMID: 27040081 Free PMC article.
References
-
- Miller BJ, Appel MC, O’Neil JJ, Wicker LS (1988) Both the Lyt-2+ and L3T4+ T cell subsets are required for the transfer of diabetes in nonobese diabetic mice. J Immunol 140: 52–58. - PubMed
-
- Tisch R, McDevitt H (1996) Insulin-dependent diabetes mellitus. Cell 85: 291–297. - PubMed
-
- Leiter EH (2005) Nonobese diabetic mice and the genetics of diabetes susceptibility. Curr Diab Rep 5: 141–148. - PubMed
-
- Anderson MS, Bluestone JA (2005) The NOD mouse: a model of immune dysregulation. Annu Rev Immunol 23: 447–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

